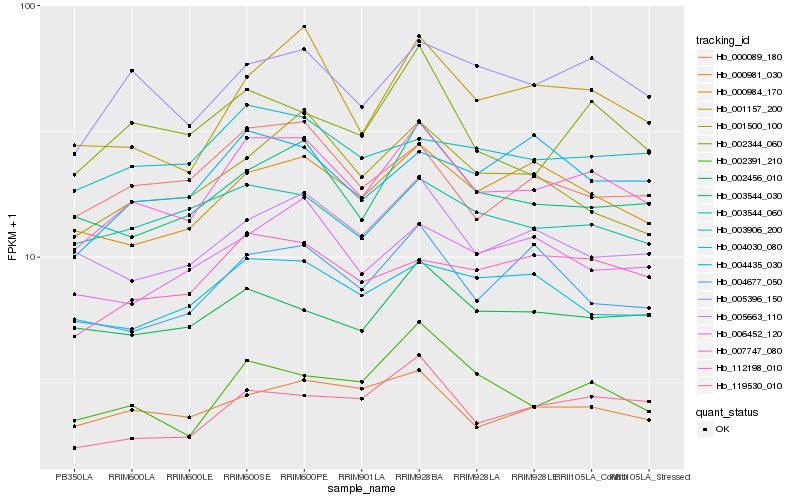
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007747_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005396_150 |
0.0738417643 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_003544_030 |
0.0761207693 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 4 |
Hb_001157_200 |
0.0784524651 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 5 |
Hb_000984_170 |
0.0818874556 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 6 |
Hb_112198_010 |
0.0824190453 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 7 |
Hb_119530_010 |
0.0835290314 |
- |
- |
- |
| 8 |
Hb_000089_180 |
0.08363016 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 9 |
Hb_000981_030 |
0.0848048416 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 10 |
Hb_005663_110 |
0.0857535016 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002344_060 |
0.0873098974 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 12 |
Hb_003544_060 |
0.0873918376 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 13 |
Hb_003906_200 |
0.0876306019 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 14 |
Hb_002391_210 |
0.0883461772 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 15 |
Hb_001500_100 |
0.0886982891 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002456_010 |
0.0903881097 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |
| 17 |
Hb_006452_120 |
0.0904749896 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 18 |
Hb_004677_050 |
0.0914158267 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004030_080 |
0.09238301 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 20 |
Hb_004435_030 |
0.0935150706 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |