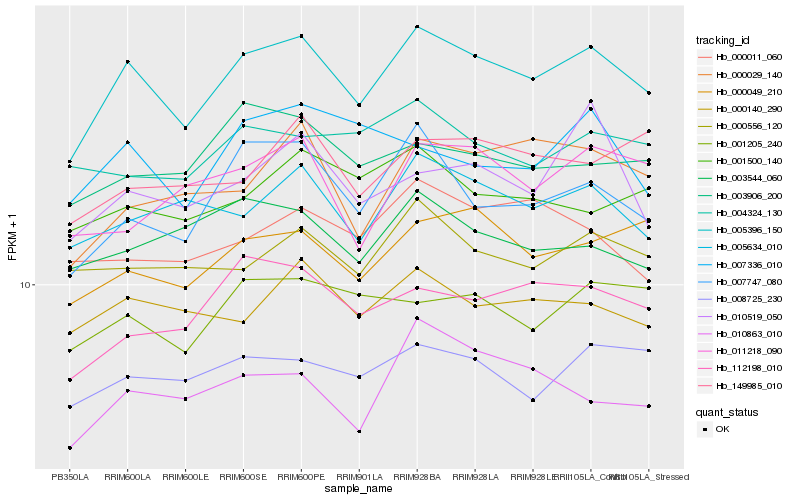
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005396_150 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_007747_080 |
0.0738417643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005634_010 |
0.0789321388 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_000049_210 |
0.0810211946 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 5 |
Hb_112198_010 |
0.0814592379 |
- |
- |
PREDICTED: F-box protein SKIP22 [Jatropha curcas] |
| 6 |
Hb_008725_230 |
0.0835285551 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |
| 7 |
Hb_000140_290 |
0.0835527923 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 59 kDa protein isoform X1 [Populus euphratica] |
| 8 |
Hb_000556_120 |
0.0858393563 |
- |
- |
Spastin, putative [Ricinus communis] |
| 9 |
Hb_001205_240 |
0.0858504041 |
- |
- |
PREDICTED: uncharacterized protein LOC105645528 [Jatropha curcas] |
| 10 |
Hb_000029_140 |
0.0878752888 |
- |
- |
PREDICTED: putative GDP-L-fucose synthase 2 [Populus euphratica] |
| 11 |
Hb_003906_200 |
0.0898987135 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 12 |
Hb_010863_010 |
0.0899731594 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_003544_060 |
0.0917220066 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 14 |
Hb_001500_140 |
0.0921728421 |
- |
- |
pelota, putative [Ricinus communis] |
| 15 |
Hb_007336_010 |
0.0924798099 |
- |
- |
PREDICTED: F-box protein At1g47056-like [Jatropha curcas] |
| 16 |
Hb_149985_010 |
0.0930985423 |
- |
- |
PREDICTED: transcription initiation factor IIB-2 [Cucumis sativus] |
| 17 |
Hb_004324_130 |
0.0934174985 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 18 |
Hb_010519_050 |
0.0937898699 |
- |
- |
galactokinase, putative [Ricinus communis] |
| 19 |
Hb_011218_090 |
0.0940017698 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 20 |
Hb_000011_060 |
0.0942988083 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |