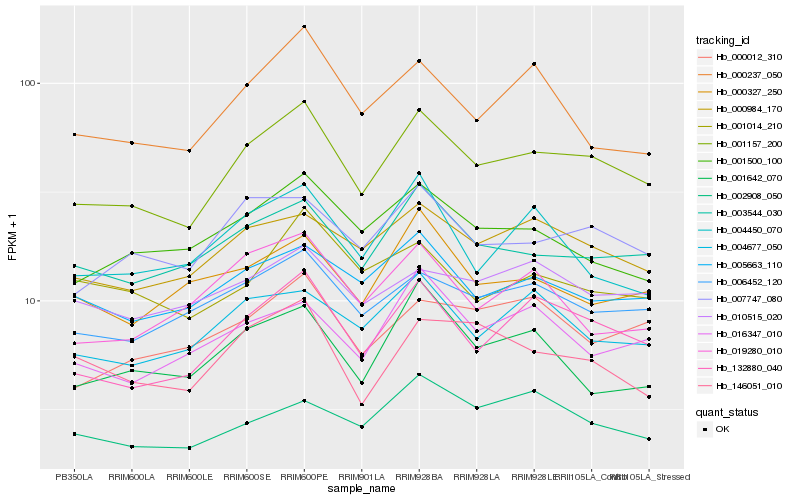
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001157_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 2 |
Hb_132880_040 |
0.0597132556 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_003544_030 |
0.0760625334 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 4 |
Hb_007747_080 |
0.0784524651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000984_170 |
0.0785100975 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 6 |
Hb_006452_120 |
0.0809416572 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 7 |
Hb_005663_110 |
0.0860509361 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 8 |
Hb_004677_050 |
0.0886000468 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001500_100 |
0.0933986451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_016347_010 |
0.0948587382 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 11 |
Hb_000012_310 |
0.0969498192 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_002908_050 |
0.097318172 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 13 |
Hb_000237_050 |
0.0976298308 |
- |
- |
CP2 [Hevea brasiliensis] |
| 14 |
Hb_146051_010 |
0.0978326642 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 15 |
Hb_010515_020 |
0.099198068 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 16 |
Hb_000327_250 |
0.1006337816 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 17 |
Hb_001014_210 |
0.1012220694 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 18 |
Hb_004450_070 |
0.1032231853 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 19 |
Hb_019280_010 |
0.1040879041 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001642_070 |
0.1042762037 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |