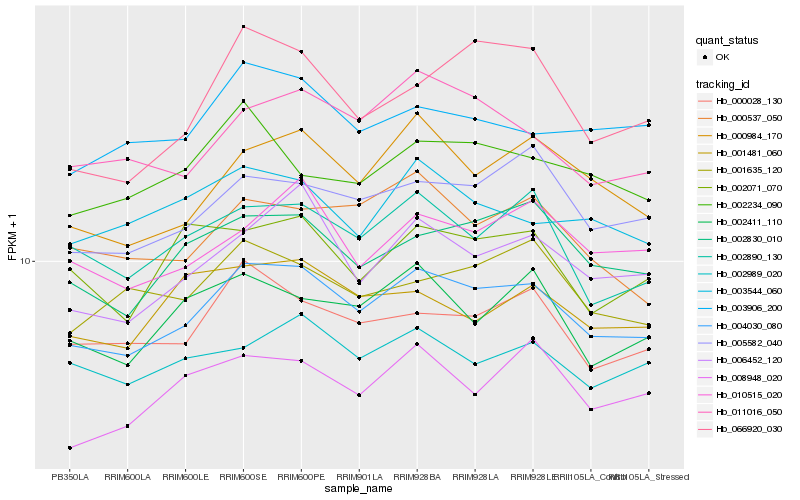
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004030_080 |
0.0 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 2 |
Hb_002830_010 |
0.0544680701 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 3 |
Hb_000984_170 |
0.0558882919 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 4 |
Hb_005582_040 |
0.0559767132 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 5 |
Hb_006452_120 |
0.0564514715 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 6 |
Hb_000028_130 |
0.0596725069 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 7 |
Hb_011016_050 |
0.0603223431 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002890_130 |
0.0615512978 |
- |
- |
tip120, putative [Ricinus communis] |
| 9 |
Hb_010515_020 |
0.0638305231 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 10 |
Hb_002411_110 |
0.0664243025 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 11 |
Hb_002989_020 |
0.0668928686 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000537_050 |
0.0670651404 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 13 |
Hb_003906_200 |
0.0673855196 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 14 |
Hb_001635_120 |
0.0683959482 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 15 |
Hb_008948_020 |
0.0688680761 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 16 |
Hb_003544_060 |
0.0691848852 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 17 |
Hb_002234_090 |
0.0698250183 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |
| 18 |
Hb_066920_030 |
0.0701911908 |
- |
- |
PREDICTED: adagio protein 1 [Jatropha curcas] |
| 19 |
Hb_002071_070 |
0.070646063 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001481_060 |
0.0708724153 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |