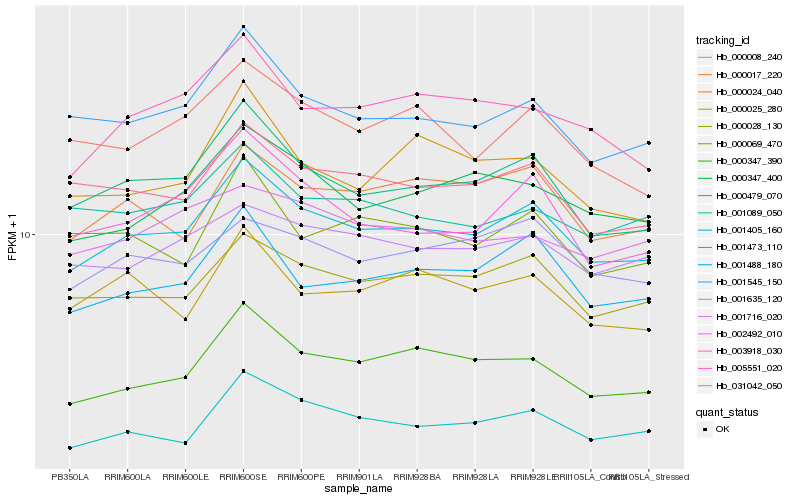
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001473_110 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 2 |
Hb_000479_070 |
0.0420564204 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 3 |
Hb_000028_130 |
0.0525714953 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001488_180 |
0.0565005432 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001545_150 |
0.057287998 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 6 |
Hb_000347_390 |
0.0602742031 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 7 |
Hb_000347_400 |
0.0613495464 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_031042_050 |
0.0615975315 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 9 |
Hb_003918_030 |
0.0617650238 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_005551_020 |
0.0667293397 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 11 |
Hb_000017_220 |
0.0668032242 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 12 |
Hb_001635_120 |
0.0674972384 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 13 |
Hb_000008_240 |
0.0681631529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001716_020 |
0.0683355172 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 15 |
Hb_000025_280 |
0.0727647992 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 16 |
Hb_001089_050 |
0.073806018 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 17 |
Hb_001405_160 |
0.0747011701 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g09680 [Jatropha curcas] |
| 18 |
Hb_000069_470 |
0.0750106771 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000024_040 |
0.0754633057 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 20 |
Hb_002492_010 |
0.0759282699 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |