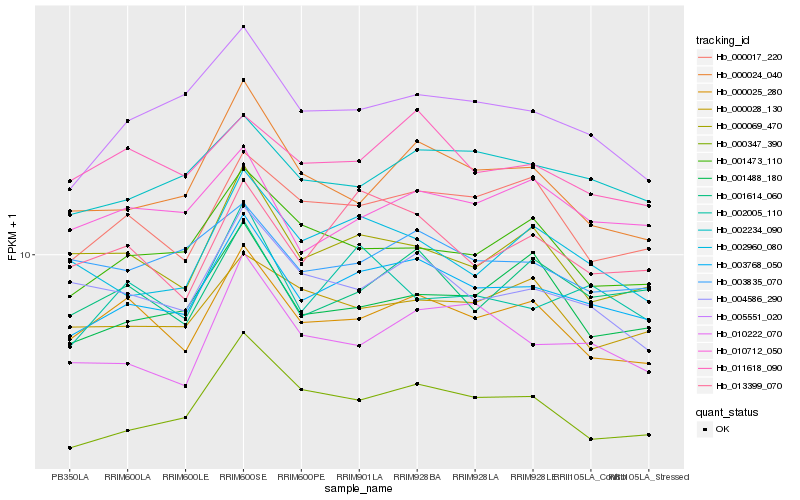
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003768_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002234_090 |
0.0634377979 |
- |
- |
PREDICTED: probable transcriptional regulator SLK2 [Jatropha curcas] |
| 3 |
Hb_000347_390 |
0.063877628 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 4 |
Hb_000025_280 |
0.0676961857 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 5 |
Hb_005551_020 |
0.0680690106 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 6 |
Hb_001614_060 |
0.0708022237 |
- |
- |
hypothetical protein JCGZ_06918 [Jatropha curcas] |
| 7 |
Hb_000017_220 |
0.0715170667 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 8 |
Hb_013399_070 |
0.0719105371 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 9 |
Hb_010222_070 |
0.0724028493 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000024_040 |
0.0746572611 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 11 |
Hb_001473_110 |
0.0761336533 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 12 |
Hb_010712_050 |
0.0761341232 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 13 |
Hb_011618_090 |
0.0769395019 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_002960_080 |
0.0779200845 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105635322 [Jatropha curcas] |
| 15 |
Hb_004586_290 |
0.077994024 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 16 |
Hb_002005_110 |
0.0783043621 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |
| 17 |
Hb_000069_470 |
0.0784632537 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001488_180 |
0.0784789161 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003835_070 |
0.078578335 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000028_130 |
0.0804309047 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |