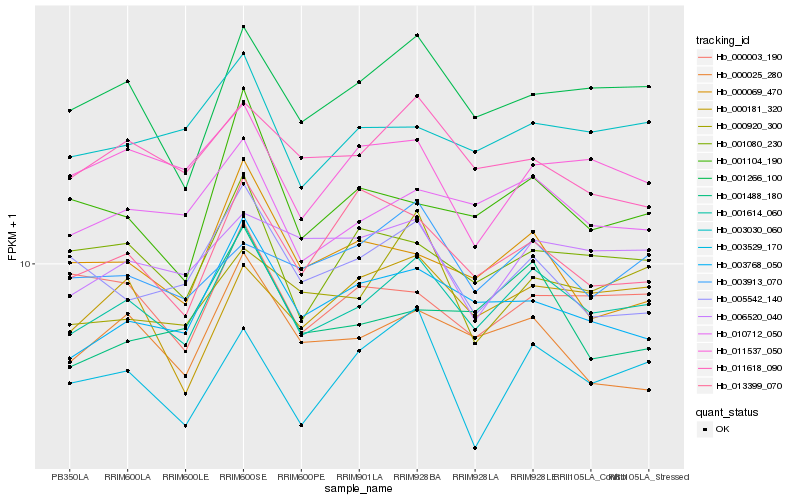
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001614_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_06918 [Jatropha curcas] |
| 2 |
Hb_010712_050 |
0.0613784987 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 3 |
Hb_001266_100 |
0.062173864 |
- |
- |
PREDICTED: uncharacterized protein LOC105649131 [Jatropha curcas] |
| 4 |
Hb_003768_050 |
0.0708022237 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001080_230 |
0.073201581 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105649979 [Jatropha curcas] |
| 6 |
Hb_003030_060 |
0.0737918113 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 7 |
Hb_000025_280 |
0.0787574627 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 8 |
Hb_000181_320 |
0.0813086586 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 9 |
Hb_000003_190 |
0.0827294552 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like isoform X1 [Nelumbo nucifera] |
| 10 |
Hb_011618_090 |
0.0830285402 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_003529_170 |
0.083043703 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX29 [Jatropha curcas] |
| 12 |
Hb_011537_050 |
0.0833115426 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001104_190 |
0.0834902605 |
- |
- |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 14 |
Hb_006520_040 |
0.0845020505 |
- |
- |
PREDICTED: uncharacterized protein LOC105641714 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000920_300 |
0.0848212951 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 16 |
Hb_013399_070 |
0.0855412021 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005542_140 |
0.0860180706 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 18 |
Hb_003913_070 |
0.0862467821 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 30 [Populus euphratica] |
| 19 |
Hb_000069_470 |
0.0862588204 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001488_180 |
0.0863419904 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |