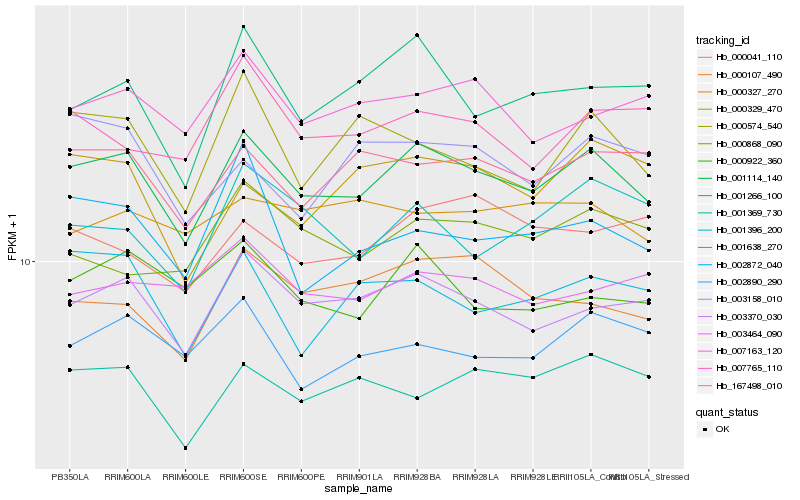
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001114_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631631 [Jatropha curcas] |
| 2 |
Hb_003370_030 |
0.0552957048 |
- |
- |
PREDICTED: uncharacterized protein LOC105635547 isoform X2 [Jatropha curcas] |
| 3 |
Hb_167498_010 |
0.0687530985 |
- |
- |
PREDICTED: WPP domain-interacting protein 1-like [Jatropha curcas] |
| 4 |
Hb_001266_100 |
0.0720829702 |
- |
- |
PREDICTED: uncharacterized protein LOC105649131 [Jatropha curcas] |
| 5 |
Hb_007163_120 |
0.0735441029 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 6 |
Hb_001369_730 |
0.0737215171 |
- |
- |
F5O11.10, putative isoform 1 [Theobroma cacao] |
| 7 |
Hb_000107_490 |
0.0742124629 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 8 |
Hb_002890_290 |
0.0758170525 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001638_270 |
0.0758938396 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 8 homolog isoform X2 [Jatropha curcas] |
| 10 |
Hb_000922_360 |
0.0762473614 |
- |
- |
- |
| 11 |
Hb_007765_110 |
0.0787678644 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 12 |
Hb_000574_540 |
0.0797951529 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_001396_200 |
0.0798169825 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000329_470 |
0.0808110311 |
- |
- |
ccr4-not transcription complex, putative [Ricinus communis] |
| 15 |
Hb_003464_090 |
0.0813452988 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 16 |
Hb_002872_040 |
0.0825702934 |
- |
- |
pumilio, putative [Ricinus communis] |
| 17 |
Hb_000041_110 |
0.0828997391 |
- |
- |
PREDICTED: WW domain-binding protein 11 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000868_090 |
0.0829080223 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 19 |
Hb_003158_010 |
0.0830912422 |
- |
- |
hypothetical protein JCGZ_17361 [Jatropha curcas] |
| 20 |
Hb_000327_270 |
0.0833219108 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |