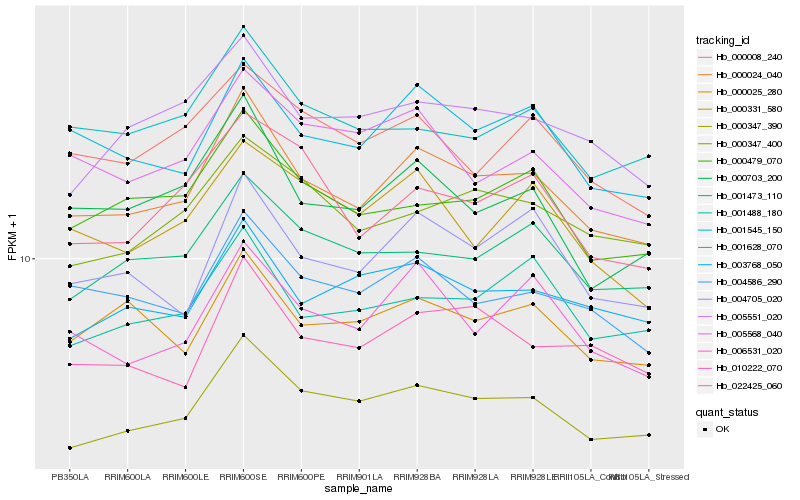
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000024_040 |
0.0 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 2 |
Hb_000347_390 |
0.0525287534 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 3 |
Hb_001628_070 |
0.0626816183 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_000703_200 |
0.0659126589 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_004586_290 |
0.0664974657 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 6 |
Hb_004705_020 |
0.0695669796 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 7 |
Hb_000479_070 |
0.0713444347 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 8 |
Hb_006531_020 |
0.072453316 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 9 |
Hb_010222_070 |
0.0725390027 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_003768_050 |
0.0746572611 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000025_280 |
0.0747884124 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 12 |
Hb_001473_110 |
0.0754633057 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |
| 13 |
Hb_005551_020 |
0.0762633691 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 14 |
Hb_001545_150 |
0.0776964015 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 15 |
Hb_001488_180 |
0.0781974101 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000347_400 |
0.0790850871 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_005568_040 |
0.0794704082 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000008_240 |
0.0795794602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_022425_060 |
0.0820527054 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 20 |
Hb_000331_580 |
0.0832012256 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |