| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
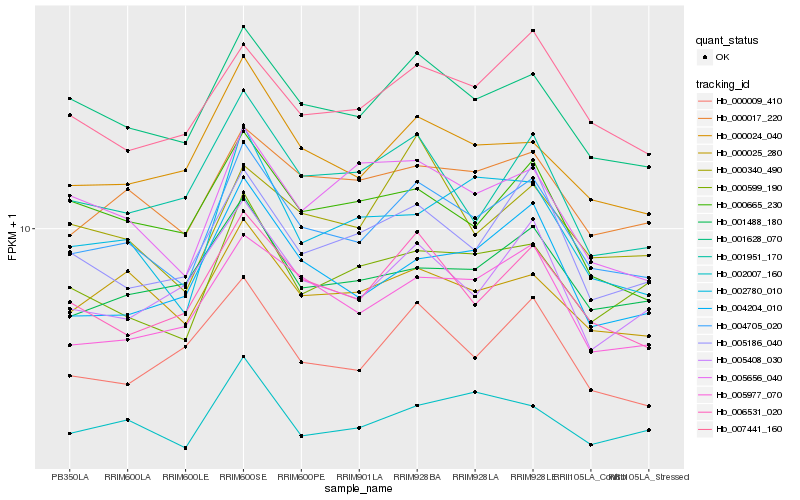
Hb_004705_020 |
0.0 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 2 |
Hb_001628_070 |
0.0540317504 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_006531_020 |
0.0641116739 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 4 |
Hb_000017_220 |
0.0672098272 |
- |
- |
PREDICTED: uncharacterized protein LOC105645768 [Jatropha curcas] |
| 5 |
Hb_000024_040 |
0.0695669796 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 6 |
Hb_000025_280 |
0.0709184147 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_07432 [Jatropha curcas] |
| 7 |
Hb_005977_070 |
0.0711506988 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005186_040 |
0.074879638 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 9 |
Hb_007441_160 |
0.0748975165 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_000599_190 |
0.0754103979 |
transcription factor |
TF Family: SNF2 |
DNA repair and recombination protein RAD26, putative [Ricinus communis] |
| 11 |
Hb_001488_180 |
0.0773053734 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004204_010 |
0.0788144225 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 13 |
Hb_002007_160 |
0.0796175958 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 14 |
Hb_005408_030 |
0.0806698077 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 15 |
Hb_001951_170 |
0.0815032476 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 16 |
Hb_000009_410 |
0.0834875203 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 17 |
Hb_002780_010 |
0.0835558081 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 18 |
Hb_000665_230 |
0.08386636 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 19 |
Hb_005656_040 |
0.0838761142 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_000340_490 |
0.0851811377 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |