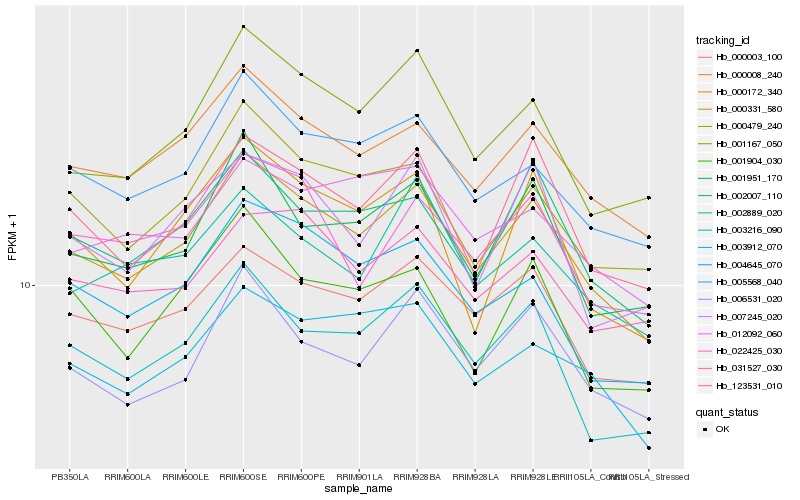
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_580 |
0.0 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 2 |
Hb_000008_240 |
0.0434560932 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000003_100 |
0.0503594872 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 4 |
Hb_005568_040 |
0.0516715073 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002007_110 |
0.0547387313 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 6 |
Hb_123531_010 |
0.055711699 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 7 |
Hb_006531_020 |
0.0572510717 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 8 |
Hb_001167_050 |
0.05763443 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 9 |
Hb_003216_090 |
0.0585964133 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001951_170 |
0.0595779754 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 11 |
Hb_003912_070 |
0.0607407476 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 12 |
Hb_000479_240 |
0.0610366909 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004645_070 |
0.0621734391 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 14 |
Hb_031527_030 |
0.0678068268 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 15 |
Hb_002889_020 |
0.0697865817 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 3 [Jatropha curcas] |
| 16 |
Hb_022425_030 |
0.0701937126 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 17 |
Hb_000172_340 |
0.0702075696 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 18 |
Hb_001904_030 |
0.0702860193 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 19 |
Hb_007245_020 |
0.0712802824 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 20 |
Hb_012092_060 |
0.0718029906 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |