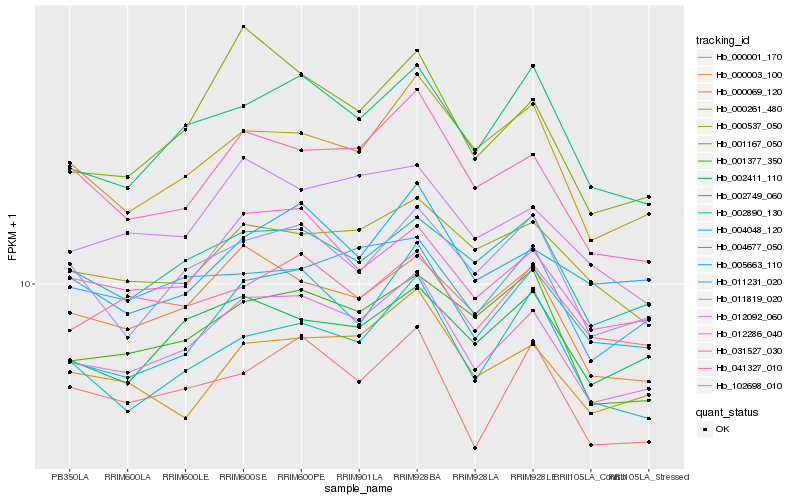
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012286_040 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 2 |
Hb_031527_030 |
0.0534003067 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 3 |
Hb_002749_060 |
0.0566921728 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 4 |
Hb_041327_010 |
0.0578915572 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 5 |
Hb_000003_100 |
0.0580103665 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 6 |
Hb_002890_130 |
0.0621801432 |
- |
- |
tip120, putative [Ricinus communis] |
| 7 |
Hb_012092_060 |
0.0629403051 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 8 |
Hb_000261_480 |
0.0647723093 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 9 |
Hb_011819_020 |
0.0648676222 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000001_170 |
0.0649386031 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_002411_110 |
0.0669731792 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |
| 12 |
Hb_102698_010 |
0.0670374398 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001377_350 |
0.0672530764 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 14 |
Hb_001167_050 |
0.0674819727 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 15 |
Hb_000069_120 |
0.0675596785 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 16 |
Hb_000537_050 |
0.0687075403 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 17 |
Hb_004048_120 |
0.0697914011 |
- |
- |
PREDICTED: nuclear pore complex protein NUP160 isoform X3 [Jatropha curcas] |
| 18 |
Hb_005663_110 |
0.0702558719 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004677_050 |
0.0706265222 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_011231_020 |
0.0719182551 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |