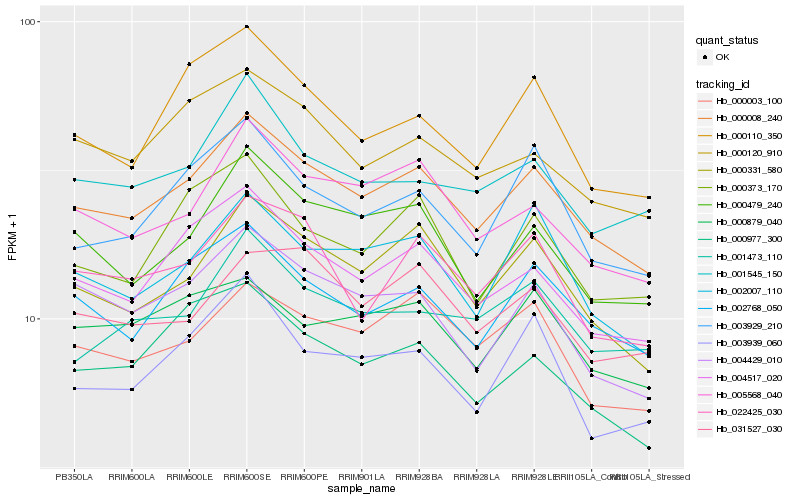
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000331_580 |
0.0434560932 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 3 |
Hb_004517_020 |
0.0489669573 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 4 |
Hb_002768_050 |
0.0519075428 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 5 |
Hb_005568_040 |
0.0536660326 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002007_110 |
0.0577756224 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000479_240 |
0.0579031332 |
- |
- |
PREDICTED: probable ubiquitin conjugation factor E4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000003_100 |
0.0591953989 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 9 |
Hb_003929_210 |
0.0616250542 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 10 |
Hb_004429_010 |
0.0620852274 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 11 |
Hb_000120_910 |
0.0625929178 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 12 |
Hb_022425_030 |
0.0630006337 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 13 |
Hb_000977_300 |
0.0659264948 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 14 |
Hb_000879_040 |
0.0660129307 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 15 |
Hb_000373_170 |
0.0662025153 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 16 |
Hb_003939_060 |
0.0663654522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000110_350 |
0.0664799341 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 18 |
Hb_001545_150 |
0.0665408726 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 19 |
Hb_031527_030 |
0.067906565 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 20 |
Hb_001473_110 |
0.0681631529 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C-like [Jatropha curcas] |