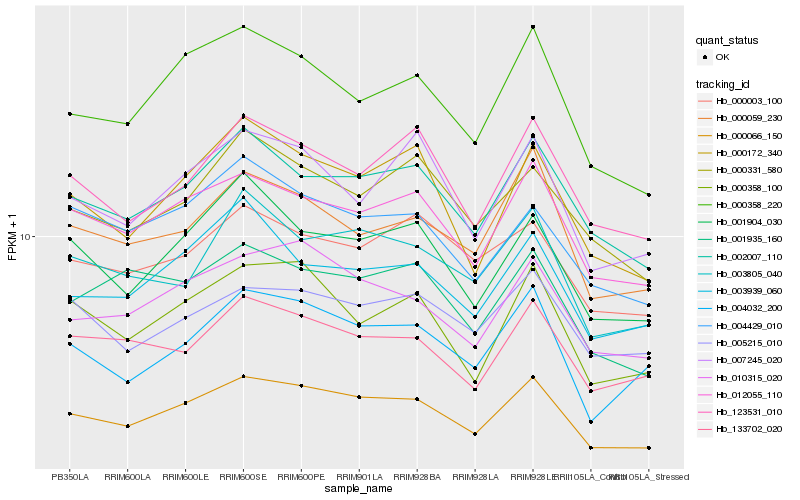
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000066_150 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 2 |
Hb_012055_110 |
0.0500914925 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 3 |
Hb_002007_110 |
0.0551600263 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 4 |
Hb_010315_020 |
0.0563795075 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 5 |
Hb_000358_220 |
0.0595570212 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 6 |
Hb_000172_340 |
0.064982908 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 7 |
Hb_004032_200 |
0.0671658367 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 8 |
Hb_001904_030 |
0.0682218965 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 9 |
Hb_000358_100 |
0.0686678845 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 10 |
Hb_000059_230 |
0.0693814317 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 11 |
Hb_123531_010 |
0.0694795778 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 12 |
Hb_133702_020 |
0.0708573836 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_003805_040 |
0.0716095128 |
- |
- |
Sacsin [Glycine soja] |
| 14 |
Hb_007245_020 |
0.0724000616 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 15 |
Hb_003939_060 |
0.0741309561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001935_160 |
0.0747784261 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 17 |
Hb_004429_010 |
0.0748712896 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 18 |
Hb_000331_580 |
0.0752309696 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 19 |
Hb_005215_010 |
0.0753486221 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000003_100 |
0.0759615902 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |