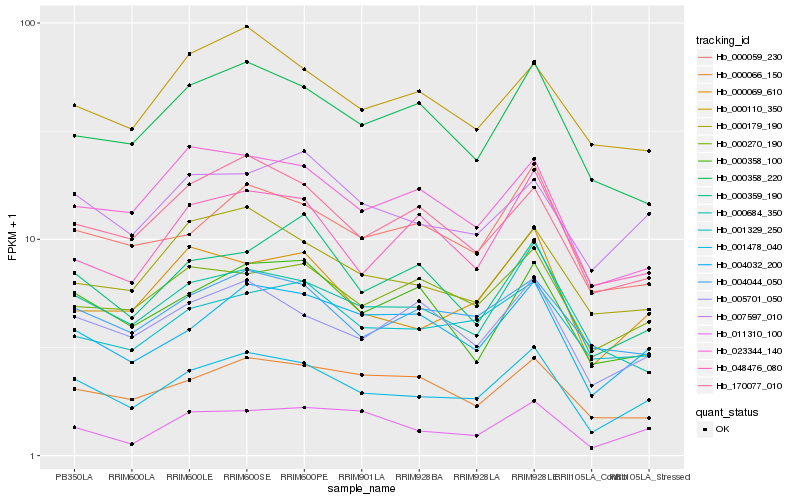
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001478_040 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like [Jatropha curcas] |
| 2 |
Hb_004032_200 |
0.0831715847 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 3 |
Hb_000069_610 |
0.0900909652 |
- |
- |
PREDICTED: protein SUPPRESSOR OF PHYA-105 1 [Jatropha curcas] |
| 4 |
Hb_005701_050 |
0.0937530333 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 5 |
Hb_000059_230 |
0.0939514409 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 6 |
Hb_007597_010 |
0.0970255428 |
- |
- |
hypothetical protein B456_010G072300 [Gossypium raimondii] |
| 7 |
Hb_170077_010 |
0.098228769 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000179_190 |
0.098865147 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 9 |
Hb_004044_050 |
0.0999194126 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 10 |
Hb_001329_250 |
0.0999871287 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000270_190 |
0.1013143331 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 12 |
Hb_000684_350 |
0.1023265736 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 13 |
Hb_023344_140 |
0.1031194286 |
- |
- |
PREDICTED: WD repeat-containing protein 70 [Jatropha curcas] |
| 14 |
Hb_000358_100 |
0.1055918343 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 15 |
Hb_048476_080 |
0.1062401722 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 16 |
Hb_011310_100 |
0.1066193743 |
- |
- |
hypothetical protein CICLE_v10008199mg [Citrus clementina] |
| 17 |
Hb_000358_220 |
0.1069031974 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 18 |
Hb_000066_150 |
0.1078695541 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 19 |
Hb_000110_350 |
0.1086393542 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Gossypium raimondii] |
| 20 |
Hb_000359_190 |
0.1086530706 |
- |
- |
PREDICTED: protein VACUOLELESS1 [Nicotiana sylvestris] |