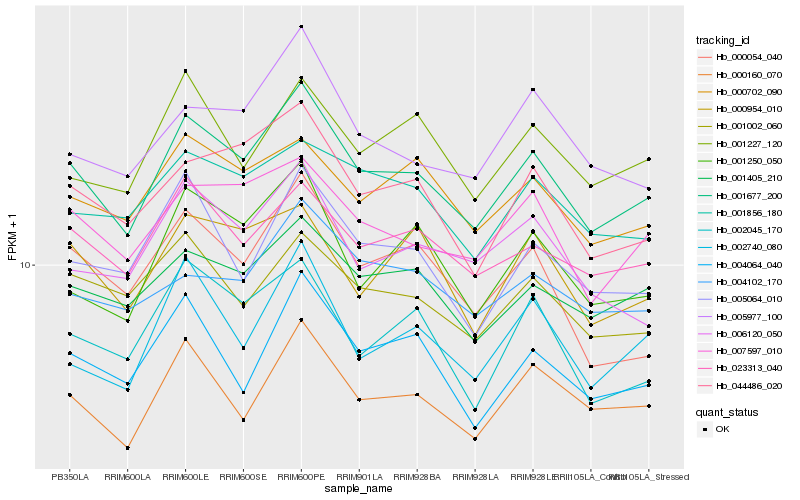
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001677_200 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_023313_040 |
0.0716656954 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001405_210 |
0.0719979576 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 4 |
Hb_007597_010 |
0.0730764901 |
- |
- |
hypothetical protein B456_010G072300 [Gossypium raimondii] |
| 5 |
Hb_044486_020 |
0.0746937248 |
- |
- |
CASTOR protein [Glycine max] |
| 6 |
Hb_000160_070 |
0.0754129191 |
- |
- |
PREDICTED: uncharacterized protein LOC105648219 [Jatropha curcas] |
| 7 |
Hb_000702_090 |
0.0811833546 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 8 |
Hb_001002_060 |
0.0815307967 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 9 |
Hb_001856_180 |
0.0838861231 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 10 |
Hb_001250_050 |
0.0840520319 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005064_010 |
0.085071923 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 12 |
Hb_005977_100 |
0.0858394552 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 13 |
Hb_001227_120 |
0.0864248815 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 14 |
Hb_004064_040 |
0.0870545899 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 15 |
Hb_006120_050 |
0.0872169102 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 16 |
Hb_004102_170 |
0.0879181478 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 17 |
Hb_000954_010 |
0.088368894 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 18 |
Hb_000054_040 |
0.0888235487 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 19 |
Hb_002740_080 |
0.0896224015 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 20 |
Hb_002045_170 |
0.0912995385 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |