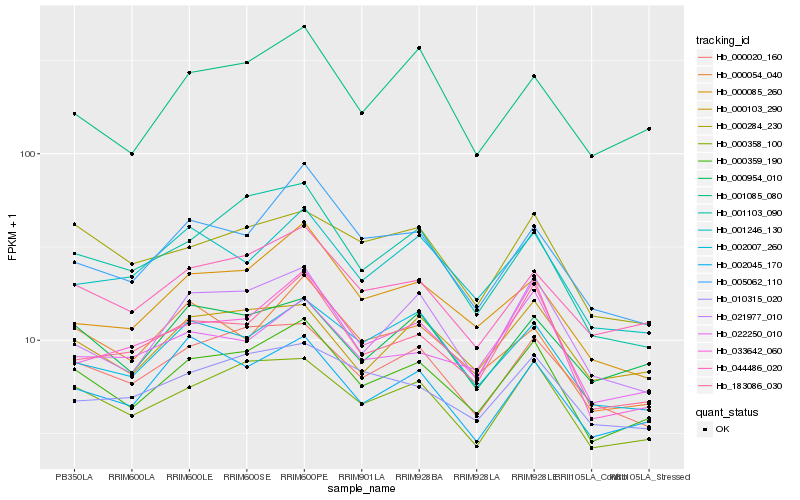
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_190 |
0.0 |
- |
- |
PREDICTED: protein VACUOLELESS1 [Nicotiana sylvestris] |
| 2 |
Hb_000358_100 |
0.0616552371 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 3 |
Hb_044486_020 |
0.0678216072 |
- |
- |
CASTOR protein [Glycine max] |
| 4 |
Hb_000020_160 |
0.0759336136 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 5 |
Hb_183086_030 |
0.0801114112 |
- |
- |
GTPase-activating protein GYP7 [Gossypium arboreum] |
| 6 |
Hb_021977_010 |
0.0837824823 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 7 |
Hb_000103_290 |
0.0839071593 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 8 |
Hb_000954_010 |
0.0848658214 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 9 |
Hb_002007_260 |
0.0848819128 |
- |
- |
beta-mannosidase, putative [Ricinus communis] |
| 10 |
Hb_000085_260 |
0.0868685543 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 11 |
Hb_000284_230 |
0.0878340553 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 12 |
Hb_000054_040 |
0.0881531668 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 13 |
Hb_033642_060 |
0.0888384901 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002045_170 |
0.0894516081 |
- |
- |
interferon-induced guanylate-binding protein, putative [Ricinus communis] |
| 15 |
Hb_001246_130 |
0.0907979108 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001085_080 |
0.0916231803 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 17 |
Hb_010315_020 |
0.0917085043 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 18 |
Hb_022250_010 |
0.0917778327 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 19 |
Hb_001103_090 |
0.0921524289 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 20 |
Hb_005062_110 |
0.0935544832 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 1 [Jatropha curcas] |