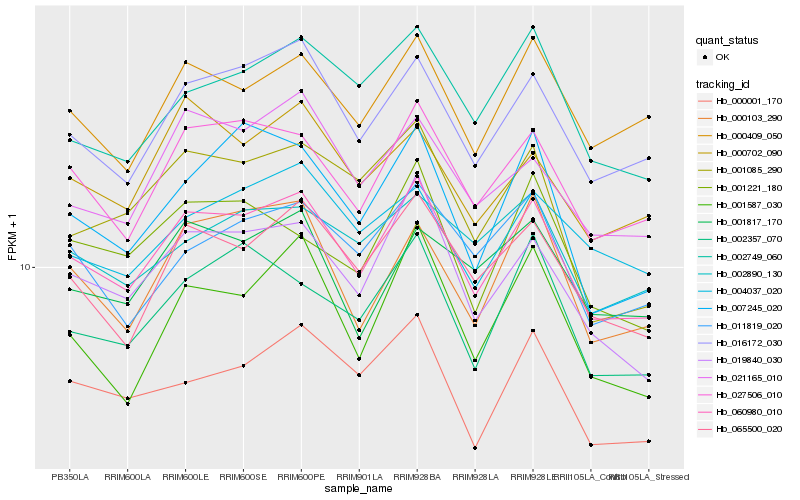
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_060980_010 |
0.0 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 2 |
Hb_016172_030 |
0.0433949501 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 3 |
Hb_027506_010 |
0.0442334508 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 4 |
Hb_019840_030 |
0.0494891729 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 5 |
Hb_065500_020 |
0.050423315 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 6 |
Hb_000409_050 |
0.0600605283 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 7 |
Hb_000001_170 |
0.0607100267 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_021165_010 |
0.061048359 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000103_290 |
0.0613728861 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 10 |
Hb_002357_070 |
0.0622772097 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 11 |
Hb_002749_060 |
0.0629415903 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 12 |
Hb_011819_020 |
0.0640000234 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007245_020 |
0.064585299 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 14 |
Hb_000702_090 |
0.065407716 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 15 |
Hb_001221_180 |
0.0671813023 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 16 |
Hb_001085_290 |
0.0673492945 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 17 |
Hb_001587_030 |
0.0678244964 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 18 |
Hb_001817_170 |
0.0682153471 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 19 |
Hb_002890_130 |
0.0684041519 |
- |
- |
tip120, putative [Ricinus communis] |
| 20 |
Hb_004037_020 |
0.0687881576 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |