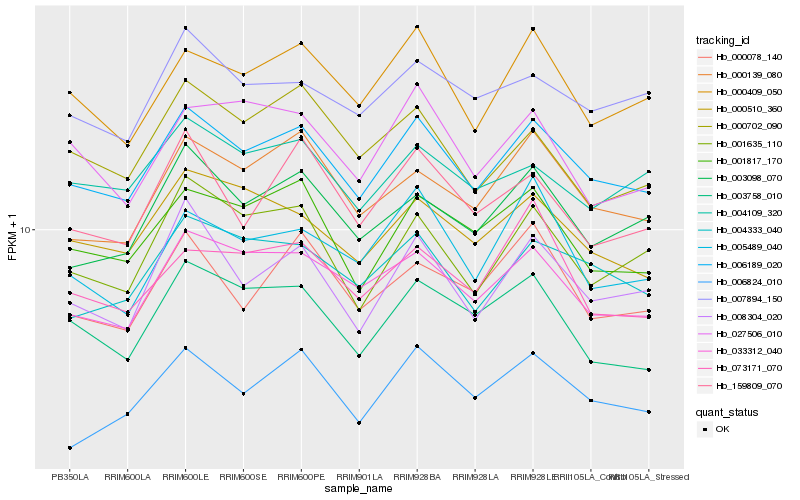
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006189_020 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 2 |
Hb_033312_040 |
0.0480705354 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000409_050 |
0.051728624 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 4 |
Hb_004333_040 |
0.0525966209 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 5 |
Hb_001817_170 |
0.0538738375 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 6 |
Hb_000510_360 |
0.0542750605 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 7 |
Hb_000139_080 |
0.0556902888 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_008304_020 |
0.056093641 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 9 |
Hb_003098_070 |
0.0571176989 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 10 |
Hb_007894_150 |
0.0597383393 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 11 |
Hb_001635_110 |
0.0603939197 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 12 |
Hb_004109_320 |
0.0621086263 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 13 |
Hb_003758_010 |
0.0631383703 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_159809_070 |
0.0665328529 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 15 |
Hb_000702_090 |
0.0674738964 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 16 |
Hb_000078_140 |
0.0679768061 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 17 |
Hb_005489_040 |
0.0680189571 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 18 |
Hb_006824_010 |
0.068964019 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 19 |
Hb_027506_010 |
0.0693013462 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 20 |
Hb_073171_070 |
0.0698441075 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |