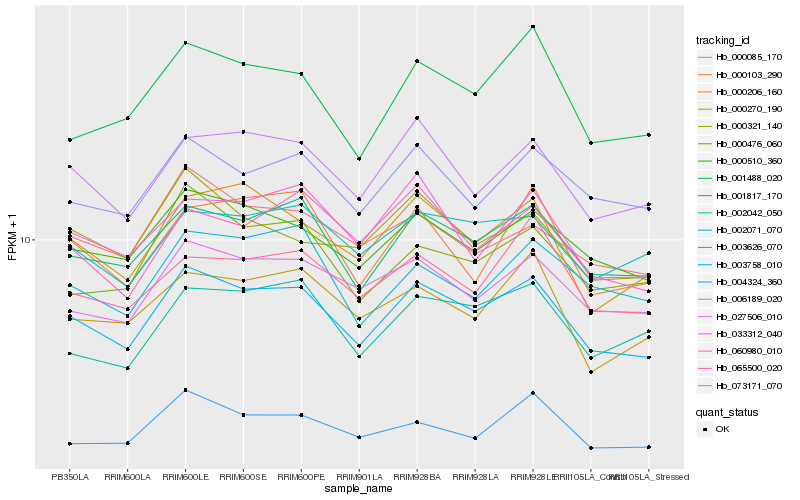
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003758_010 |
0.0 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001817_170 |
0.0468744969 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 3 |
Hb_000510_360 |
0.0521408219 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 4 |
Hb_027506_010 |
0.0589880327 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 5 |
Hb_033312_040 |
0.0610618853 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000206_160 |
0.0627536227 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_006189_020 |
0.0631383703 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 8 |
Hb_002071_070 |
0.0669150916 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001488_020 |
0.0680030148 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 10 |
Hb_060980_010 |
0.0690731315 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 11 |
Hb_000085_170 |
0.0703226648 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_073171_070 |
0.0715010132 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 13 |
Hb_000270_190 |
0.0717945261 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 14 |
Hb_002042_050 |
0.0722640016 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004324_360 |
0.0723354517 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000321_140 |
0.0724202828 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 17 |
Hb_065500_020 |
0.0726342785 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 18 |
Hb_000476_060 |
0.0730296825 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_003626_070 |
0.0754873141 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 20 |
Hb_000103_290 |
0.0755665256 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |