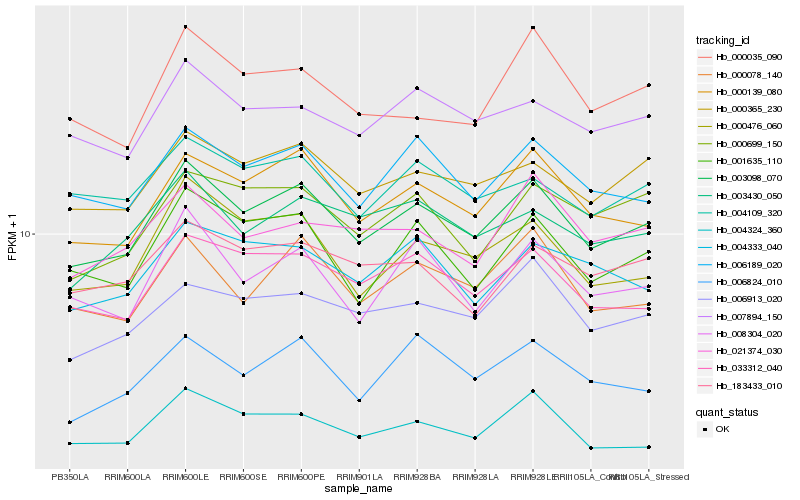
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003098_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 2 |
Hb_000139_080 |
0.0472905079 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000365_230 |
0.0561294186 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 4 |
Hb_006189_020 |
0.0571176989 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 5 |
Hb_033312_040 |
0.0580042736 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000476_060 |
0.0590737322 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_000078_140 |
0.0603273167 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 8 |
Hb_001635_110 |
0.0607594911 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 9 |
Hb_008304_020 |
0.0639183231 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 10 |
Hb_006913_020 |
0.0641636835 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003430_050 |
0.0648304075 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |
| 12 |
Hb_007894_150 |
0.0651907757 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 13 |
Hb_004324_360 |
0.0678166015 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000699_150 |
0.0692769859 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 15 |
Hb_004333_040 |
0.0701744936 |
- |
- |
PREDICTED: uncharacterized protein LOC105629988 [Jatropha curcas] |
| 16 |
Hb_004109_320 |
0.0708173677 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 17 |
Hb_183433_010 |
0.0710268649 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 18 |
Hb_006824_010 |
0.0711940126 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 19 |
Hb_021374_030 |
0.0713561379 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 20 |
Hb_000035_090 |
0.0714066474 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |