| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
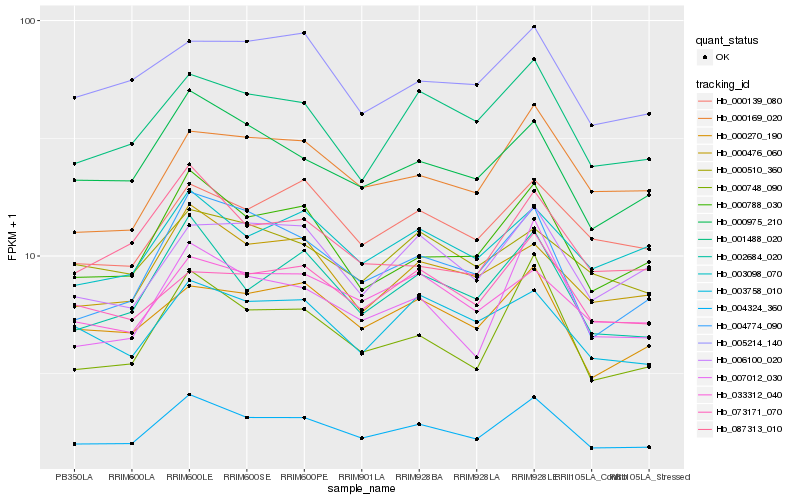
Hb_004324_360 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002684_020 |
0.0588189294 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 3 |
Hb_000975_210 |
0.0601334681 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 4 |
Hb_000788_030 |
0.0614124835 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 5 |
Hb_001488_020 |
0.0644706238 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 6 |
Hb_000476_060 |
0.0654919558 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_033312_040 |
0.0657170958 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000270_190 |
0.067351936 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 9 |
Hb_000139_080 |
0.0674671503 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003098_070 |
0.0678166015 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 11 |
Hb_000748_090 |
0.0698192061 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 12 |
Hb_087313_010 |
0.070546148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004774_090 |
0.0709648167 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 14 |
Hb_000169_020 |
0.071163082 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 15 |
Hb_003758_010 |
0.0723354517 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_073171_070 |
0.0728733366 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 17 |
Hb_006100_020 |
0.0748092307 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 18 |
Hb_000510_360 |
0.0752569524 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 19 |
Hb_005214_140 |
0.0764628102 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 20 |
Hb_007012_030 |
0.0776151322 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |