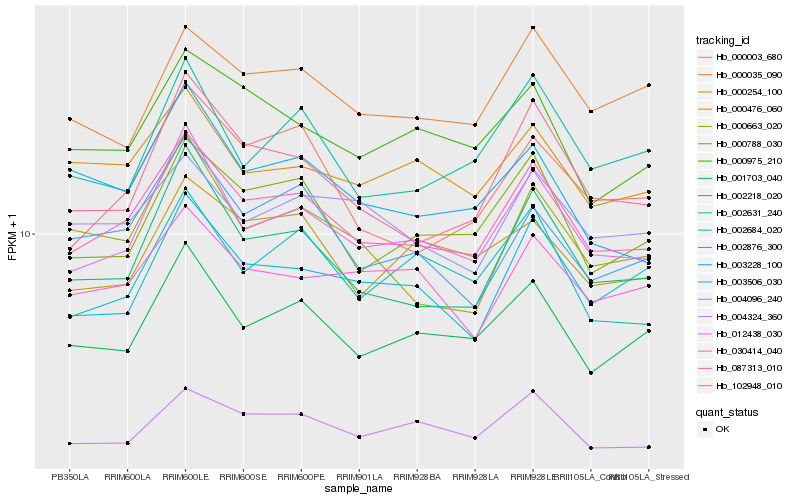
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_087313_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012438_030 |
0.0509432762 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 3 |
Hb_002218_020 |
0.0664550772 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 4 |
Hb_001703_040 |
0.0676749124 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000788_030 |
0.0676814434 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 6 |
Hb_102948_010 |
0.0681891952 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 7 |
Hb_004324_360 |
0.070546148 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000975_210 |
0.0752827096 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 9 |
Hb_000254_100 |
0.0787044668 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 10 |
Hb_003506_030 |
0.0791264224 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 11 |
Hb_002876_300 |
0.0794944934 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_004096_240 |
0.0799725018 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 13 |
Hb_000003_680 |
0.0806138958 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 14 |
Hb_030414_040 |
0.0807415153 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000035_090 |
0.0816384698 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 16 |
Hb_000663_020 |
0.0846754658 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_002631_240 |
0.0848257523 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 18 |
Hb_000476_060 |
0.085026179 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_003228_100 |
0.0851992998 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 20 |
Hb_002684_020 |
0.0853471643 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |