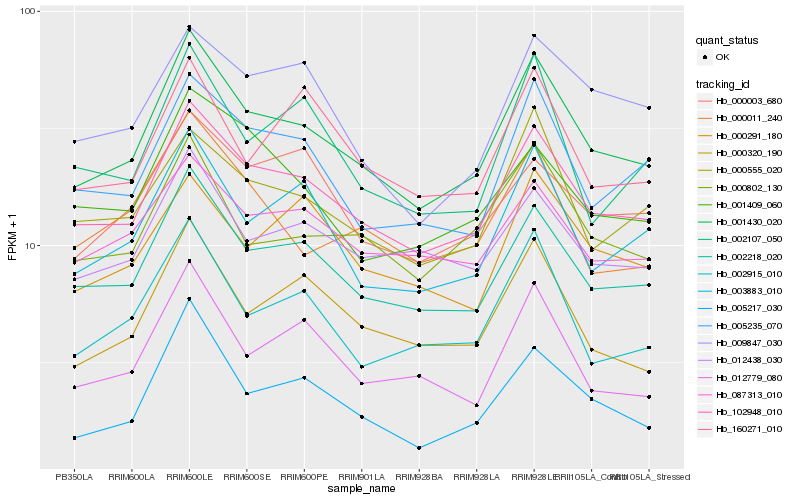
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001430_020 |
0.0 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_102948_010 |
0.0410293425 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002218_020 |
0.0567948215 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 4 |
Hb_000802_130 |
0.0829062365 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 5 |
Hb_003883_010 |
0.0848974131 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 6 |
Hb_005235_070 |
0.0863692538 |
- |
- |
PREDICTED: plastid lipid-associated protein 3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000555_020 |
0.0882776524 |
- |
- |
APO protein 2, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_000320_190 |
0.0883180267 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_002915_010 |
0.0888089028 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 10 |
Hb_012438_030 |
0.0897314773 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 11 |
Hb_087313_010 |
0.0935566907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000011_240 |
0.0965095419 |
- |
- |
PREDICTED: uncharacterized protein LOC105631316 [Jatropha curcas] |
| 13 |
Hb_012779_080 |
0.0988448861 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 14 |
Hb_005217_030 |
0.0995444367 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 15 |
Hb_000003_680 |
0.1010927322 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 16 |
Hb_000291_180 |
0.1016875828 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 17 |
Hb_009847_030 |
0.101846761 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 18 |
Hb_002107_050 |
0.1021786196 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 19 |
Hb_160271_010 |
0.1029463714 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_001409_060 |
0.1034217381 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |