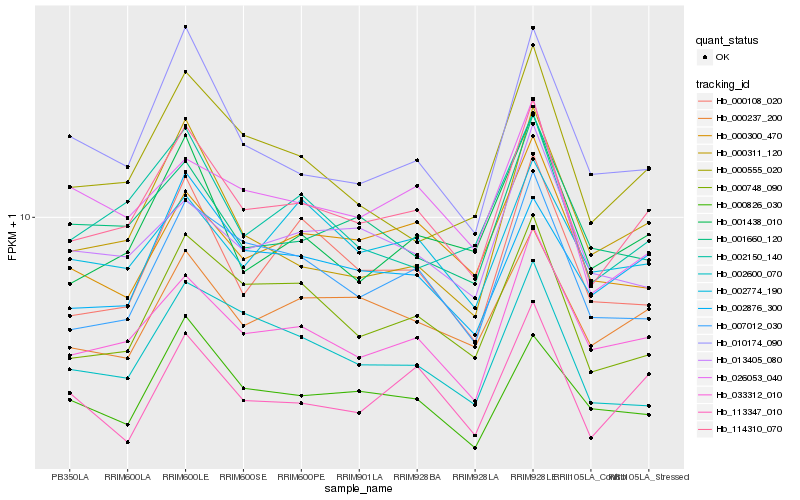
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114310_070 |
0.0 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |
| 2 |
Hb_033312_010 |
0.0632069752 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 3 |
Hb_000237_200 |
0.075933774 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 4 |
Hb_000748_090 |
0.0782819908 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 5 |
Hb_007012_030 |
0.0799049559 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002876_300 |
0.0820919357 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_002150_140 |
0.082420695 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 8 |
Hb_001438_010 |
0.0844960311 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000555_020 |
0.0848870157 |
- |
- |
APO protein 2, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_000108_020 |
0.0886109184 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 11 |
Hb_010174_090 |
0.0901123517 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001660_120 |
0.0913294707 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000300_470 |
0.0921957401 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 14 |
Hb_002774_190 |
0.0924480092 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 15 |
Hb_002600_070 |
0.0928582466 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 16 |
Hb_013405_080 |
0.0929190322 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 17 |
Hb_113347_010 |
0.0930739875 |
- |
- |
hypothetical protein CISIN_1g009336mg [Citrus sinensis] |
| 18 |
Hb_026053_040 |
0.0931864165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000826_030 |
0.0935522882 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase isoform X1 [Jatropha curcas] |
| 20 |
Hb_000311_120 |
0.0952166159 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |