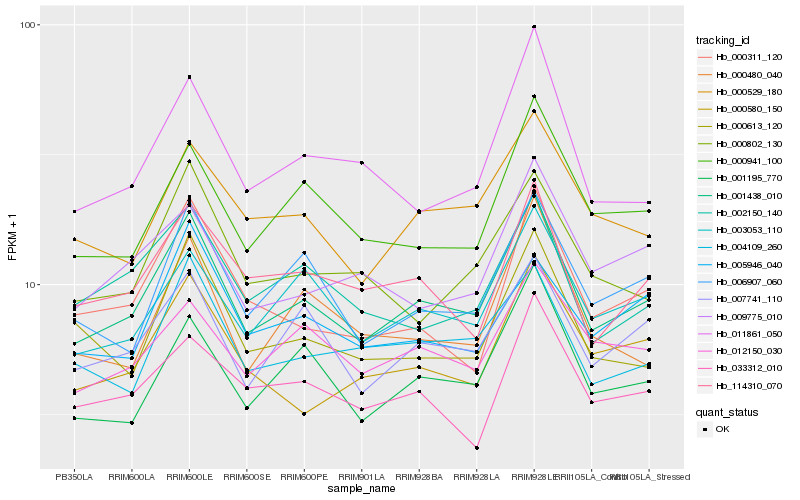
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001438_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007741_110 |
0.0744014372 |
- |
- |
- |
| 3 |
Hb_003053_110 |
0.077238986 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001195_770 |
0.078528016 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 5 |
Hb_006907_060 |
0.0803970865 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_000941_100 |
0.0813446621 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 7 |
Hb_000529_180 |
0.0823283893 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_114310_070 |
0.0844960311 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |
| 9 |
Hb_004109_260 |
0.086074083 |
- |
- |
hypothetical protein JCGZ_06843 [Jatropha curcas] |
| 10 |
Hb_012150_030 |
0.0867482767 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_002150_140 |
0.0883372011 |
- |
- |
PREDICTED: girdin-like [Jatropha curcas] |
| 12 |
Hb_000580_150 |
0.0896352833 |
- |
- |
PREDICTED: probable tRNA N6-adenosine threonylcarbamoyltransferase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_011861_050 |
0.0912530939 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_033312_010 |
0.0922030446 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_000613_120 |
0.0930041429 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000311_120 |
0.0934324485 |
- |
- |
PREDICTED: NADPH-dependent thioredoxin reductase 3 [Populus euphratica] |
| 17 |
Hb_009775_010 |
0.093952196 |
- |
- |
PREDICTED: uncharacterized protein LOC105633555 [Jatropha curcas] |
| 18 |
Hb_000480_040 |
0.0954008572 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 19 |
Hb_000802_130 |
0.0959724184 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 20 |
Hb_005946_040 |
0.0963688284 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |