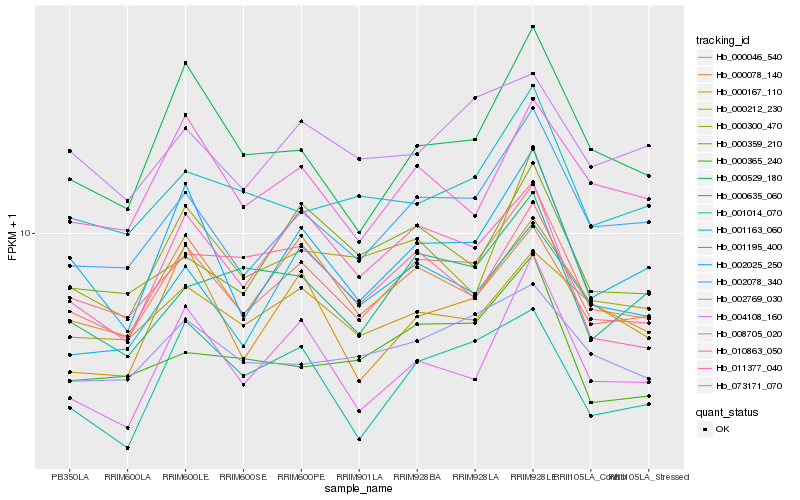
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_540 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 2 |
Hb_002078_340 |
0.0531616251 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002025_250 |
0.0672631223 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 4 |
Hb_000078_140 |
0.0766240029 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 5 |
Hb_001195_400 |
0.0768848503 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 6 |
Hb_002769_030 |
0.0774953759 |
- |
- |
PREDICTED: MATE efflux family protein 4, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000300_470 |
0.0792567722 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 8 |
Hb_010863_050 |
0.0806019739 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 9 |
Hb_000359_210 |
0.0807086314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008705_020 |
0.0817958381 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_000212_230 |
0.0825993039 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 12 |
Hb_000529_180 |
0.0826234917 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001163_060 |
0.0827842579 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 14 |
Hb_000635_060 |
0.0838863585 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 15 |
Hb_073171_070 |
0.0846926587 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 16 |
Hb_000365_240 |
0.0851345949 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33A-like [Jatropha curcas] |
| 17 |
Hb_001014_070 |
0.0852677989 |
- |
- |
PREDICTED: uncharacterized protein LOC105647305 [Jatropha curcas] |
| 18 |
Hb_004108_160 |
0.0863355453 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 19 |
Hb_011377_040 |
0.0869058498 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 20 |
Hb_000167_110 |
0.0877352063 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |