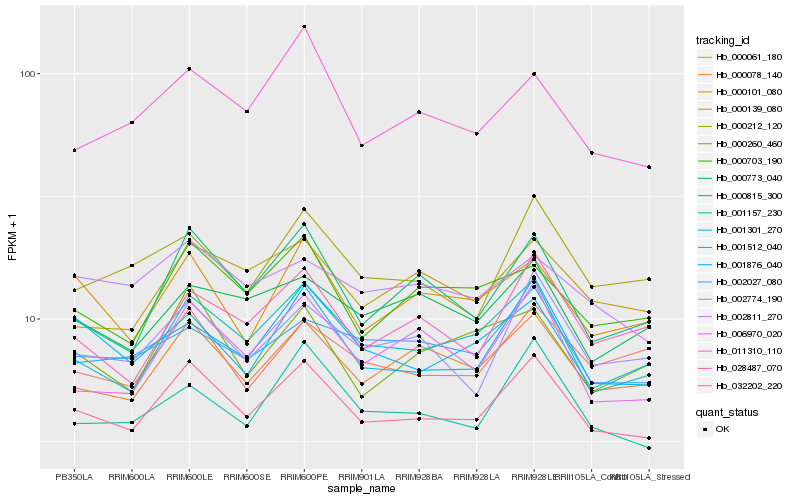
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032202_220 |
0.0 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 2 |
Hb_001301_270 |
0.0584571604 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 3 |
Hb_000078_140 |
0.0602803397 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 4 |
Hb_001876_040 |
0.0647016916 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 5 |
Hb_002774_190 |
0.0710356346 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 6 |
Hb_001157_230 |
0.0717342056 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 7 |
Hb_000061_180 |
0.0739457202 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 8 |
Hb_001512_040 |
0.0742736194 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 9 |
Hb_000815_300 |
0.0750468248 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 10 |
Hb_000101_080 |
0.0754742736 |
- |
- |
PREDICTED: protein CREG1 [Jatropha curcas] |
| 11 |
Hb_000703_190 |
0.0755770264 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002027_080 |
0.0763237349 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 13 |
Hb_000139_080 |
0.0765139006 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000212_120 |
0.0767083034 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 15 |
Hb_028487_070 |
0.0769767609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006970_020 |
0.0805710956 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 17 |
Hb_002811_270 |
0.0806941545 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 18 |
Hb_000260_460 |
0.0814262192 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000773_040 |
0.0820839231 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 20 |
Hb_011310_110 |
0.0828664145 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |