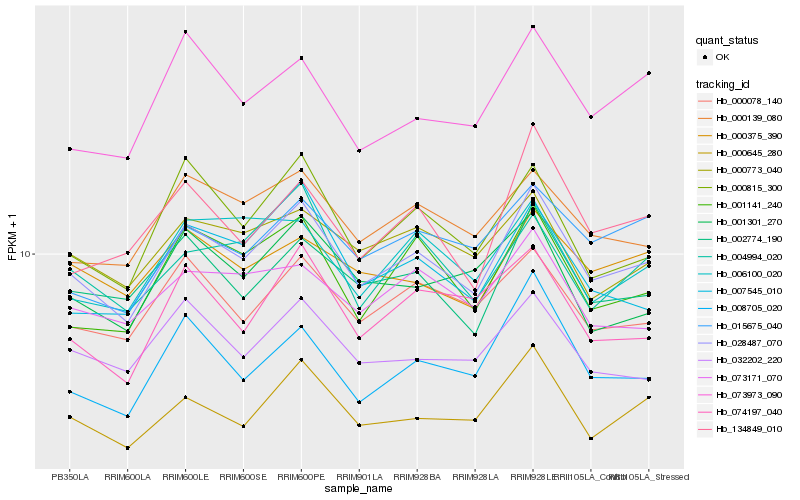
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028487_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000645_280 |
0.0669874106 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 3 |
Hb_002774_190 |
0.0683944436 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 4 |
Hb_000078_140 |
0.0701417056 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 5 |
Hb_000139_080 |
0.0717127435 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000815_300 |
0.075343036 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 7 |
Hb_001141_240 |
0.0760693446 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_032202_220 |
0.0769767609 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 9 |
Hb_000773_040 |
0.0775630619 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000375_390 |
0.0804831153 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 11 |
Hb_073171_070 |
0.0814836303 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 12 |
Hb_007545_010 |
0.08167343 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 13 |
Hb_073973_090 |
0.0817241802 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 14 |
Hb_134849_010 |
0.0818952576 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 15 |
Hb_008705_020 |
0.0821378893 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_015675_040 |
0.0821654292 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 17 |
Hb_074197_040 |
0.082344416 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 18 |
Hb_006100_020 |
0.0846838742 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 19 |
Hb_001301_270 |
0.0847249597 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 20 |
Hb_004994_020 |
0.084938313 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |