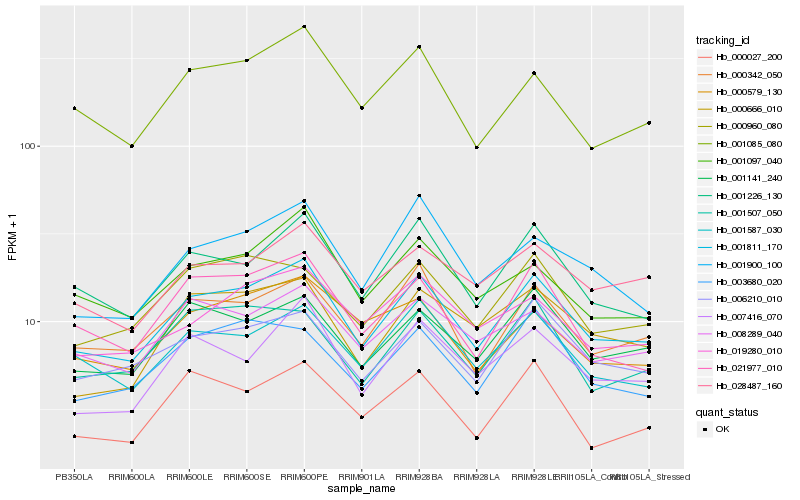
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001811_170 |
0.0 |
- |
- |
dynamin, putative [Ricinus communis] |
| 2 |
Hb_001226_130 |
0.0657080449 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 3 |
Hb_000342_050 |
0.0697116839 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |
| 4 |
Hb_001141_240 |
0.0716654843 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_006210_010 |
0.0718980414 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |
| 6 |
Hb_001587_030 |
0.0727973824 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 7 |
Hb_001085_080 |
0.078381845 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 8 |
Hb_021977_010 |
0.0784100558 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 9 |
Hb_008289_040 |
0.0790467159 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 10 |
Hb_000960_080 |
0.0811832055 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_003680_020 |
0.0815870057 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_007416_070 |
0.0823319984 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 13 |
Hb_028487_160 |
0.0835283951 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 14 |
Hb_001900_100 |
0.0840419139 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001097_040 |
0.0841849101 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 16 |
Hb_001507_050 |
0.0847454373 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_019280_010 |
0.0862547842 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000666_010 |
0.0863103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000027_200 |
0.0874975428 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 20 |
Hb_000579_130 |
0.0875536535 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |