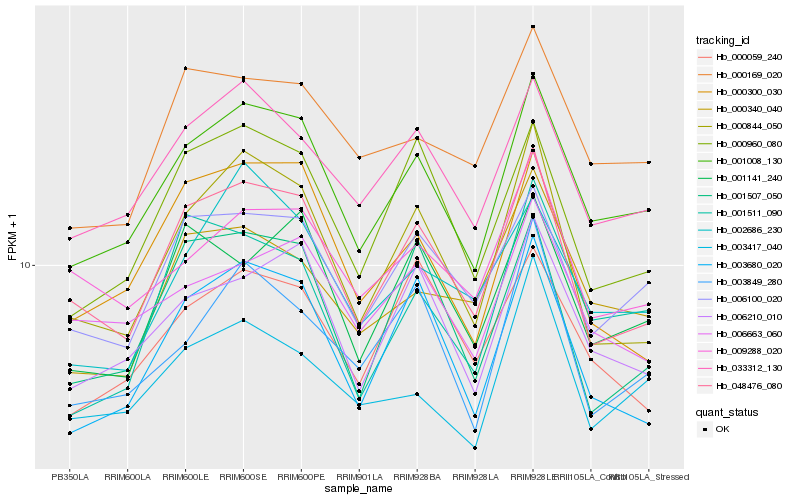
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001008_130 |
0.0 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 2 |
Hb_033312_130 |
0.054191011 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 3 |
Hb_006210_010 |
0.0575047833 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |
| 4 |
Hb_006100_020 |
0.0618920344 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 5 |
Hb_048476_080 |
0.0621007977 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 6 |
Hb_003680_020 |
0.0682859762 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000960_080 |
0.0684394338 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000169_020 |
0.0724191495 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 9 |
Hb_001141_240 |
0.0756279189 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000844_050 |
0.0764821586 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 11 |
Hb_003849_280 |
0.0765079855 |
- |
- |
PREDICTED: uncharacterized protein LOC105644977 [Jatropha curcas] |
| 12 |
Hb_002686_230 |
0.0773849668 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_003417_040 |
0.0773911723 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 14 |
Hb_006663_060 |
0.0786291181 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 15 |
Hb_001511_090 |
0.0795185541 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 16 |
Hb_000340_040 |
0.0797572123 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000300_030 |
0.0802114992 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 18 |
Hb_000059_240 |
0.0813069005 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 19 |
Hb_009288_020 |
0.081886626 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 20 |
Hb_001507_050 |
0.0820583453 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |