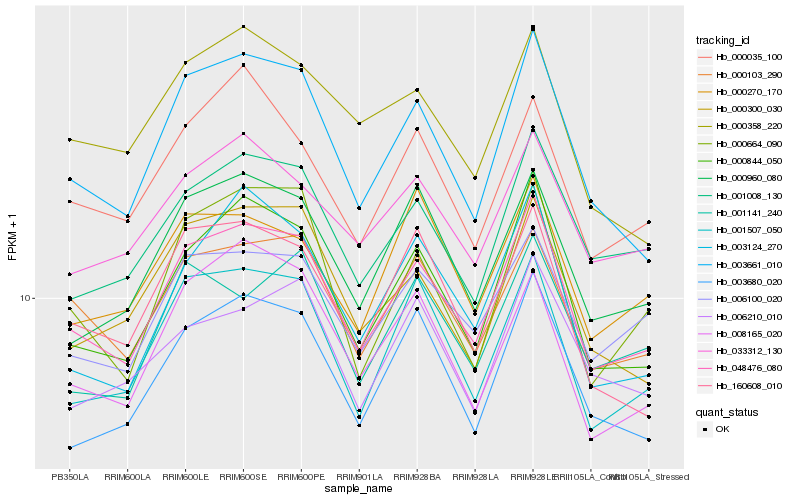
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048476_080 |
0.0 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 2 |
Hb_000844_050 |
0.045800605 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 3 |
Hb_003661_010 |
0.0508351234 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000103_290 |
0.055846152 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 5 |
Hb_001507_050 |
0.0579933075 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_006100_020 |
0.0617432893 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 7 |
Hb_001008_130 |
0.0621007977 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 8 |
Hb_003680_020 |
0.0625382868 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000960_080 |
0.0644085063 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_033312_130 |
0.0673312681 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000300_030 |
0.0686082166 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 12 |
Hb_160608_010 |
0.0699660165 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 13 |
Hb_000035_100 |
0.0709199853 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_008165_020 |
0.0713978605 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003124_270 |
0.0739420887 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 16 |
Hb_000664_090 |
0.0741162681 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 17 |
Hb_001141_240 |
0.0753405086 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000270_170 |
0.0757027102 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 19 |
Hb_006210_010 |
0.0763584839 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |
| 20 |
Hb_000358_220 |
0.0766264114 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |