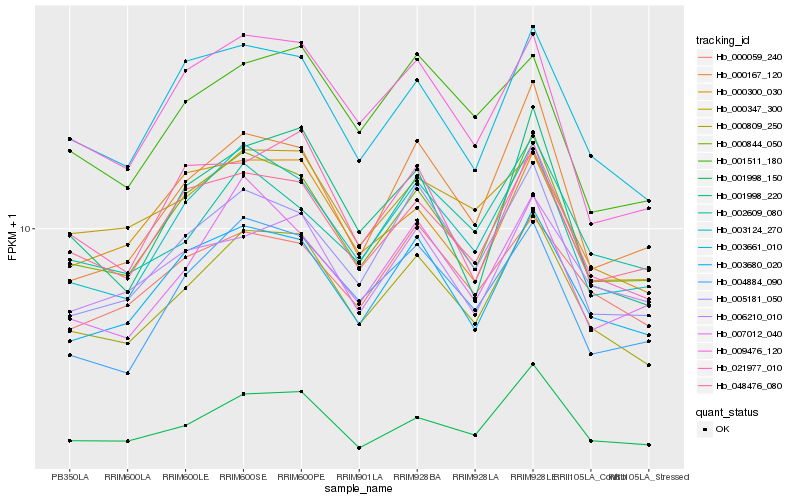
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000809_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 2 |
Hb_000844_050 |
0.0556522014 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 3 |
Hb_003124_270 |
0.0625370141 |
transcription factor |
TF Family: LUG |
hypothetical protein JCGZ_13223 [Jatropha curcas] |
| 4 |
Hb_003680_020 |
0.0659299183 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_001998_150 |
0.0707271043 |
- |
- |
hypothetical protein JCGZ_09140 [Jatropha curcas] |
| 6 |
Hb_003661_010 |
0.0714740721 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005181_050 |
0.0734931919 |
- |
- |
PREDICTED: phytochrome B isoform X1 [Jatropha curcas] |
| 8 |
Hb_004884_090 |
0.0776879819 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 9 |
Hb_048476_080 |
0.0791081483 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 10 |
Hb_009476_120 |
0.0829238554 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 11 |
Hb_001998_220 |
0.0834758084 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000167_120 |
0.0873941144 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 13 |
Hb_006210_010 |
0.0882598568 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |
| 14 |
Hb_007012_040 |
0.0889449699 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 15 |
Hb_000059_240 |
0.0893019721 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 16 |
Hb_000347_300 |
0.0894878794 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 17 |
Hb_000300_030 |
0.0904663433 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 18 |
Hb_021977_010 |
0.090634945 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 19 |
Hb_002609_080 |
0.0916120337 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_001511_180 |
0.091921122 |
- |
- |
Clathrin heavy chain 2 -like protein [Gossypium arboreum] |