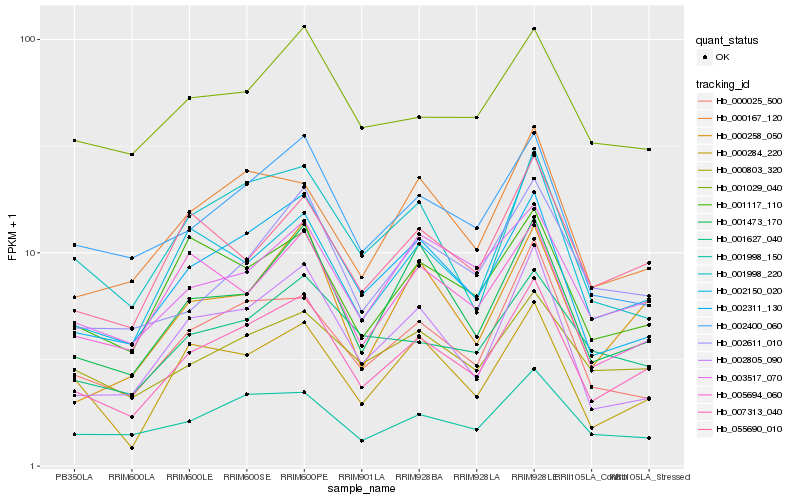
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007313_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 2 |
Hb_002311_130 |
0.0792627971 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001998_150 |
0.0927014393 |
- |
- |
hypothetical protein JCGZ_09140 [Jatropha curcas] |
| 4 |
Hb_001473_170 |
0.0958491865 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 5 |
Hb_000167_120 |
0.0985197037 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 6 |
Hb_003517_070 |
0.1023135108 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 7 |
Hb_000025_500 |
0.1036859983 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 8 |
Hb_000803_320 |
0.1038296127 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 9 |
Hb_002805_090 |
0.1038783782 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 10 |
Hb_001117_110 |
0.1049379706 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 11 |
Hb_002611_010 |
0.1071762429 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_055690_010 |
0.1073540143 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002400_060 |
0.1083809389 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 14 |
Hb_001627_040 |
0.108650597 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000258_050 |
0.1088283575 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002150_020 |
0.1091001801 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_001029_040 |
0.1107537612 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 18 |
Hb_005694_060 |
0.1110414719 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 19 |
Hb_001998_220 |
0.1110593621 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000284_220 |
0.1113538706 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |