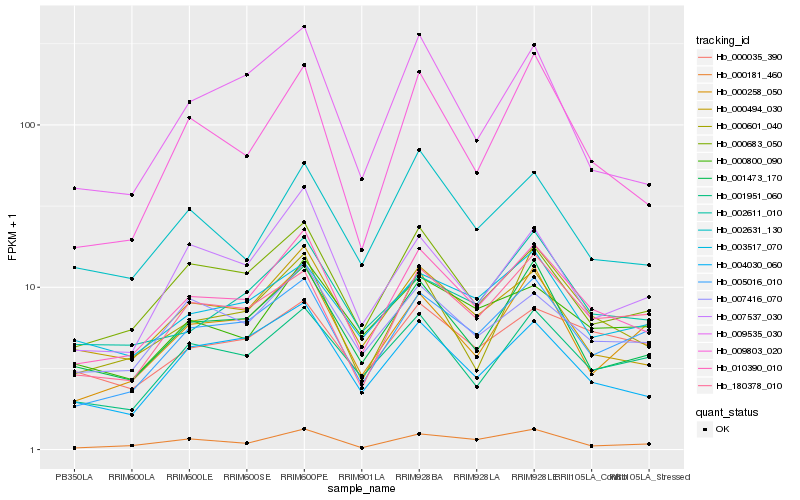
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010390_010 |
0.0 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 2 |
Hb_000494_030 |
0.0870410566 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 3 |
Hb_004030_060 |
0.0893808737 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 4 |
Hb_000683_050 |
0.0897313137 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 5 |
Hb_005016_010 |
0.0911296094 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 6 |
Hb_001473_170 |
0.0918553727 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 7 |
Hb_007416_070 |
0.0936247786 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 8 |
Hb_001951_060 |
0.1023669871 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 9 |
Hb_000601_040 |
0.1030761042 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BOI-like [Jatropha curcas] |
| 10 |
Hb_180378_010 |
0.1033566743 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 11 |
Hb_002631_130 |
0.1085346565 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 12 |
Hb_000800_090 |
0.1089430112 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000181_460 |
0.1095764886 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 14 |
Hb_007537_030 |
0.1129038534 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003517_070 |
0.1136128817 |
- |
- |
PREDICTED: protein transport protein Sec24-like At3g07100 [Jatropha curcas] |
| 16 |
Hb_002611_010 |
0.1139422329 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_000258_050 |
0.1170919619 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000035_390 |
0.1176076179 |
- |
- |
hypothetical protein CISIN_1g003355mg [Citrus sinensis] |
| 19 |
Hb_009535_030 |
0.1176767815 |
- |
- |
CP2 [Hevea brasiliensis] |
| 20 |
Hb_009803_020 |
0.1178746662 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |