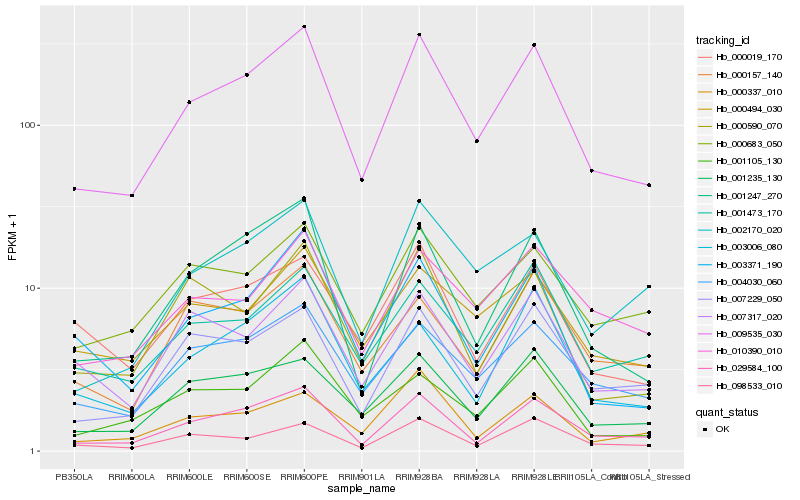
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009535_030 |
0.0 |
- |
- |
CP2 [Hevea brasiliensis] |
| 2 |
Hb_029584_100 |
0.0851590989 |
- |
- |
Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase [Gossypium arboreum] |
| 3 |
Hb_004030_060 |
0.0879752368 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 4 |
Hb_001235_130 |
0.0913503577 |
- |
- |
- |
| 5 |
Hb_001247_270 |
0.1019436045 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_007229_050 |
0.1019602654 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001473_170 |
0.1045801454 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 8 |
Hb_098533_010 |
0.1115885564 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 9 |
Hb_000683_050 |
0.1124229859 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 10 |
Hb_007317_020 |
0.1138233826 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001105_130 |
0.114576802 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 12 |
Hb_000494_030 |
0.1147490289 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 13 |
Hb_010390_010 |
0.1176767815 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 14 |
Hb_000019_170 |
0.1183777978 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 15 |
Hb_003371_190 |
0.1189456107 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 16 |
Hb_002170_020 |
0.1194536282 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |
| 17 |
Hb_003006_080 |
0.1206894796 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000157_140 |
0.1219283264 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 19 |
Hb_000337_010 |
0.1221637226 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000590_070 |
0.1226855616 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |