| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
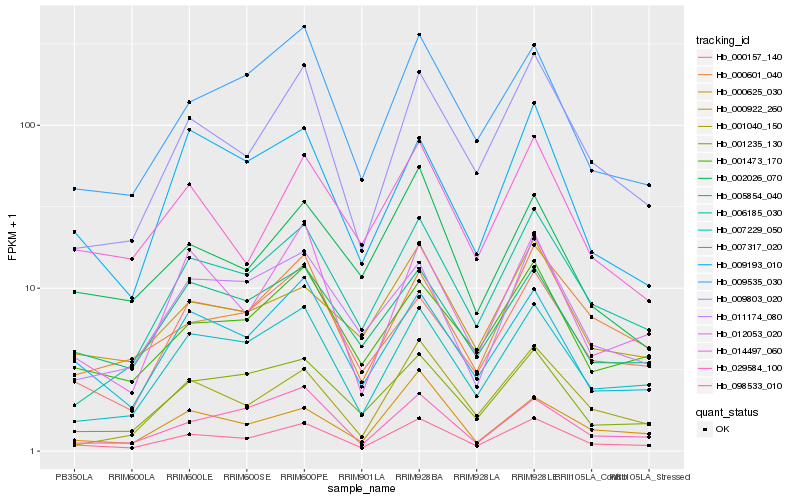
Hb_098533_010 |
0.0 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 2 |
Hb_009803_020 |
0.0874201116 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 3 |
Hb_007229_050 |
0.0889124442 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007317_020 |
0.1005680381 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006185_030 |
0.1058778863 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 6 |
Hb_001473_170 |
0.108635362 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 7 |
Hb_009535_030 |
0.1115885564 |
- |
- |
CP2 [Hevea brasiliensis] |
| 8 |
Hb_012053_020 |
0.1130545491 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001235_130 |
0.1154144117 |
- |
- |
- |
| 10 |
Hb_001040_150 |
0.1159522712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000922_260 |
0.1195842191 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |
| 12 |
Hb_029584_100 |
0.1218680813 |
- |
- |
Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase [Gossypium arboreum] |
| 13 |
Hb_000601_040 |
0.1223216092 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BOI-like [Jatropha curcas] |
| 14 |
Hb_014497_060 |
0.125082803 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 15 |
Hb_000157_140 |
0.1254879404 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 16 |
Hb_005854_040 |
0.1269959693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000625_030 |
0.1279441256 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 18 |
Hb_009193_010 |
0.1295861345 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 19 |
Hb_011174_080 |
0.130093878 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 20 |
Hb_002026_070 |
0.1304155094 |
- |
- |
hypothetical protein PHAVU_002G2910001g, partial [Phaseolus vulgaris] |