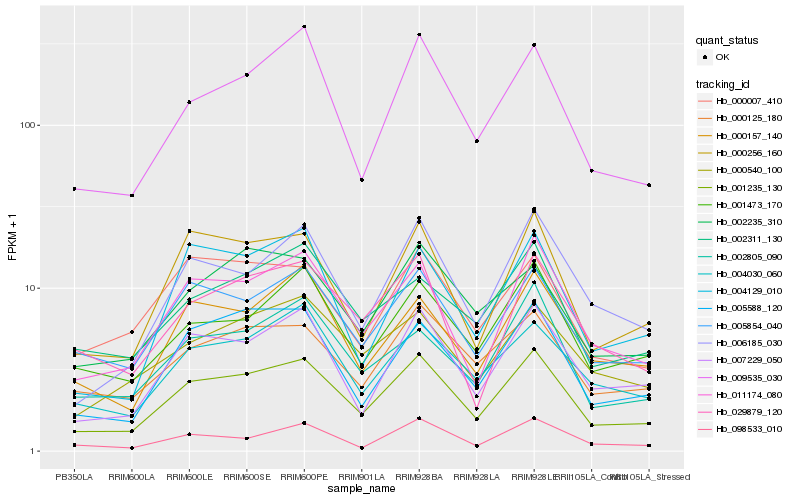
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001235_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_011174_080 |
0.0783099438 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 3 |
Hb_000256_160 |
0.0829717197 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 4 |
Hb_007229_050 |
0.0857820482 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005854_040 |
0.0875574779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005588_120 |
0.0905781421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009535_030 |
0.0913503577 |
- |
- |
CP2 [Hevea brasiliensis] |
| 8 |
Hb_004030_060 |
0.096477157 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 9 |
Hb_006185_030 |
0.097530656 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 10 |
Hb_000125_180 |
0.0982345067 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 11 |
Hb_001473_170 |
0.0984946965 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 12 |
Hb_000157_140 |
0.1005868541 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 13 |
Hb_002805_090 |
0.1034606211 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 14 |
Hb_000540_100 |
0.1070723328 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g60050 [Jatropha curcas] |
| 15 |
Hb_004129_010 |
0.1090761356 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 16 |
Hb_029879_120 |
0.1110724886 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002311_130 |
0.1115865676 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002235_310 |
0.1125854599 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 19 |
Hb_000007_410 |
0.1150147398 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |
| 20 |
Hb_098533_010 |
0.1154144117 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |