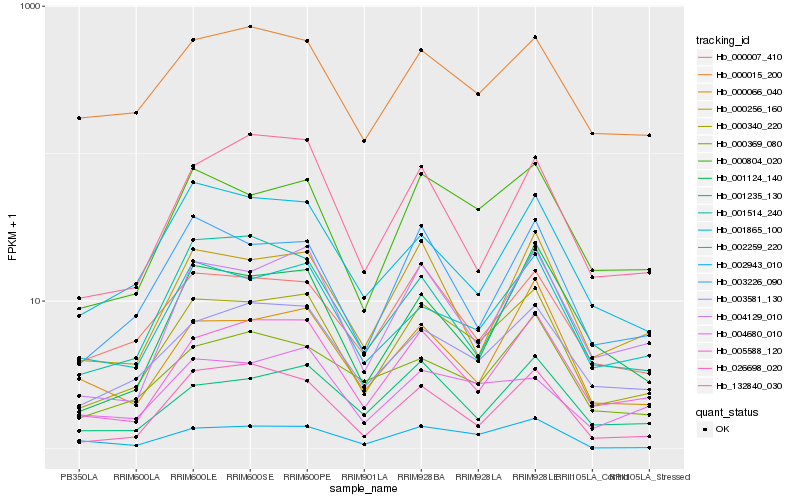
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_220 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000804_020 |
0.0866849411 |
- |
- |
PREDICTED: aconitate hydratase, cytoplasmic [Jatropha curcas] |
| 3 |
Hb_003581_130 |
0.0951033063 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_005588_120 |
0.0957541263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004129_010 |
0.1085800493 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 6 |
Hb_001865_100 |
0.1106747758 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 7 |
Hb_000007_410 |
0.1126101591 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |
| 8 |
Hb_003226_090 |
0.1129063815 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 9 |
Hb_026698_020 |
0.1211876027 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 10 |
Hb_000256_160 |
0.1215069172 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 11 |
Hb_000015_200 |
0.121879723 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: heat shock cognate 70 kDa protein 2-like [Jatropha curcas] |
| 12 |
Hb_001235_130 |
0.1248440832 |
- |
- |
- |
| 13 |
Hb_002259_220 |
0.131687722 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 14 |
Hb_000066_040 |
0.13279668 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 15 |
Hb_002943_010 |
0.1341266731 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 16 |
Hb_001514_240 |
0.1341310847 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 17 |
Hb_000369_080 |
0.1341800156 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 18 |
Hb_132840_030 |
0.1344103513 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 19 |
Hb_004680_010 |
0.1357543409 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001124_140 |
0.1374604269 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |