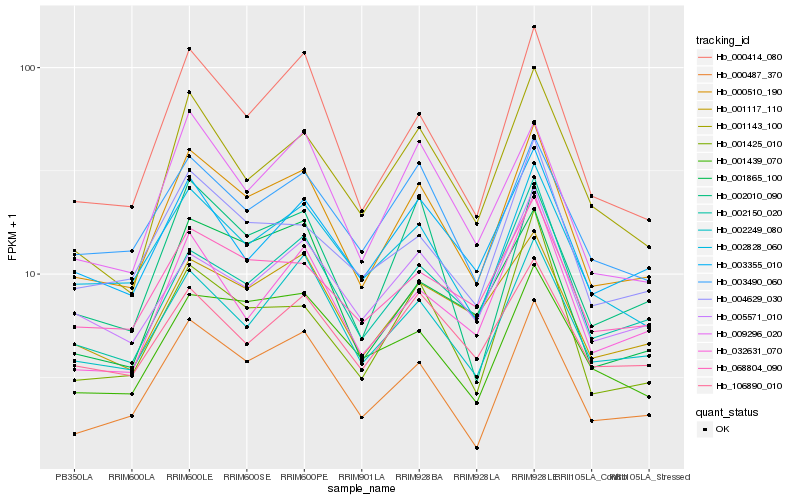
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000510_190 |
0.0 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 2 |
Hb_000487_370 |
0.0708785033 |
- |
- |
- |
| 3 |
Hb_000414_080 |
0.0734281558 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002828_060 |
0.0794156276 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002249_080 |
0.0806538969 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 6 |
Hb_009296_020 |
0.0845119007 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_003490_060 |
0.0845641194 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 8 |
Hb_001117_110 |
0.0882631028 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 9 |
Hb_001143_100 |
0.0886473924 |
- |
- |
PREDICTED: delta-aminolevulinic acid dehydratase, chloroplastic [Populus euphratica] |
| 10 |
Hb_068804_090 |
0.0894294433 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 11 |
Hb_004629_030 |
0.0906003412 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_003355_010 |
0.09243685 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 13 |
Hb_001425_010 |
0.0943010956 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 14 |
Hb_002150_020 |
0.0945189052 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_106890_010 |
0.0951042874 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_002010_090 |
0.0956674306 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_001439_070 |
0.0959984381 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_005571_010 |
0.0963104803 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 19 |
Hb_001865_100 |
0.0963842989 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 20 |
Hb_032631_070 |
0.0967151098 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |