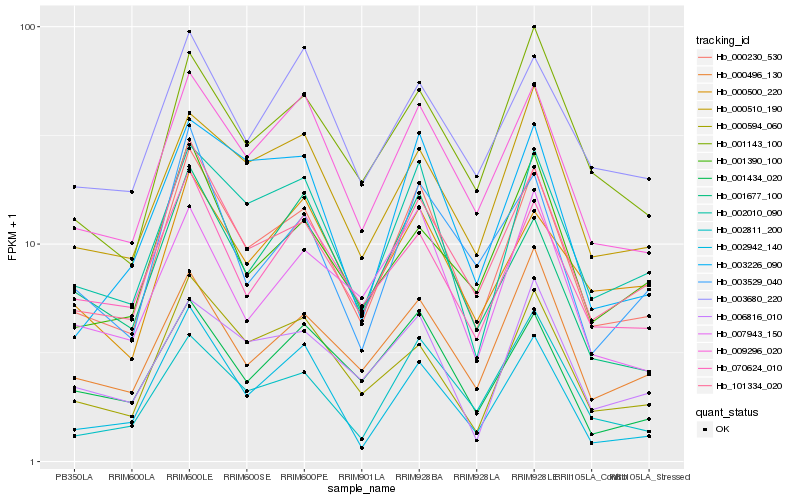
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_530 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_101334_020 |
0.0657176418 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 3 |
Hb_009296_020 |
0.0750016752 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_002010_090 |
0.0764669515 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000594_060 |
0.1015242789 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 6 |
Hb_000510_190 |
0.1028382175 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 7 |
Hb_000496_130 |
0.1034660013 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 8 |
Hb_070624_010 |
0.1064173126 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 9 |
Hb_001434_020 |
0.1064719183 |
- |
- |
hypothetical protein RCOM_0841800 [Ricinus communis] |
| 10 |
Hb_003226_090 |
0.1076962118 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 11 |
Hb_001143_100 |
0.1093650207 |
- |
- |
PREDICTED: delta-aminolevulinic acid dehydratase, chloroplastic [Populus euphratica] |
| 12 |
Hb_002811_200 |
0.1115342731 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14-like [Jatropha curcas] |
| 13 |
Hb_001390_100 |
0.1132222314 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003529_040 |
0.1134075932 |
- |
- |
PREDICTED: ATP phosphoribosyltransferase 2, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_006816_010 |
0.1136247175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002942_140 |
0.1139937013 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 17 |
Hb_007943_150 |
0.114127781 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_003680_220 |
0.1148043791 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 19 |
Hb_000500_220 |
0.1151797874 |
- |
- |
PREDICTED: intersectin-1 [Jatropha curcas] |
| 20 |
Hb_001677_100 |
0.1153688259 |
- |
- |
delta1-pyrroline-5-carboxylate synthase [Manihot esculenta] |