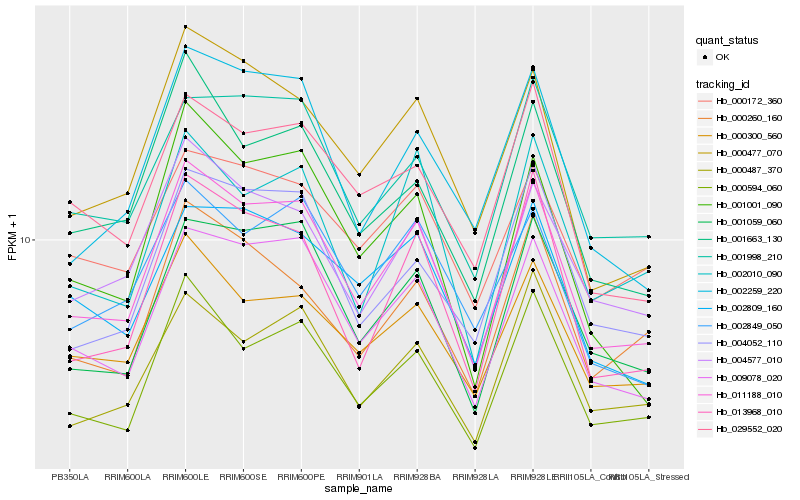
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011188_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 2 |
Hb_009078_020 |
0.0549801953 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 3 |
Hb_001059_060 |
0.0736122871 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 4 |
Hb_000300_560 |
0.0761773374 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000477_070 |
0.0764617984 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 6 |
Hb_000594_060 |
0.0768261307 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 7 |
Hb_004577_010 |
0.0786749507 |
- |
- |
PREDICTED: transmembrane protein 45A [Prunus mume] |
| 8 |
Hb_002259_220 |
0.0833290091 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 9 |
Hb_013968_010 |
0.0873946786 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 10 |
Hb_001998_210 |
0.0885536403 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_004052_110 |
0.0885568857 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_029552_020 |
0.0886311296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002849_050 |
0.0896587344 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 14 |
Hb_001663_130 |
0.0903408302 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 15 |
Hb_002809_160 |
0.093328974 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 16 |
Hb_000260_160 |
0.0962367424 |
transcription factor |
TF Family: GNAT |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_001001_090 |
0.0971101093 |
- |
- |
PREDICTED: uncharacterized protein LOC105633658 [Jatropha curcas] |
| 18 |
Hb_002010_090 |
0.0971982278 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000487_370 |
0.0982631417 |
- |
- |
- |
| 20 |
Hb_000172_360 |
0.099217065 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |