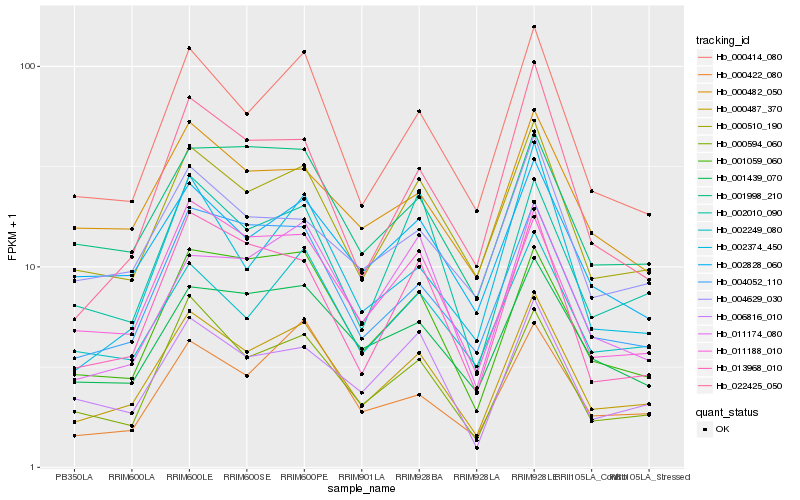
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000487_370 |
0.0 |
- |
- |
- |
| 2 |
Hb_000414_080 |
0.0660304612 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000510_190 |
0.0708785033 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 4 |
Hb_000594_060 |
0.0835849961 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 5 |
Hb_004052_110 |
0.0924850253 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001059_060 |
0.0959096882 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 7 |
Hb_002249_080 |
0.0976900362 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 8 |
Hb_011188_010 |
0.0982631417 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001439_070 |
0.098966067 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_000422_080 |
0.0989910854 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 11 |
Hb_006816_010 |
0.0992723883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002828_060 |
0.1029849933 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002010_090 |
0.1041123101 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_013968_010 |
0.1070081818 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 15 |
Hb_022425_050 |
0.1072883222 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 16 |
Hb_001998_210 |
0.1086599633 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_002374_450 |
0.1097006613 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004629_030 |
0.1119927487 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_000482_050 |
0.1129951812 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_011174_080 |
0.1136714982 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |