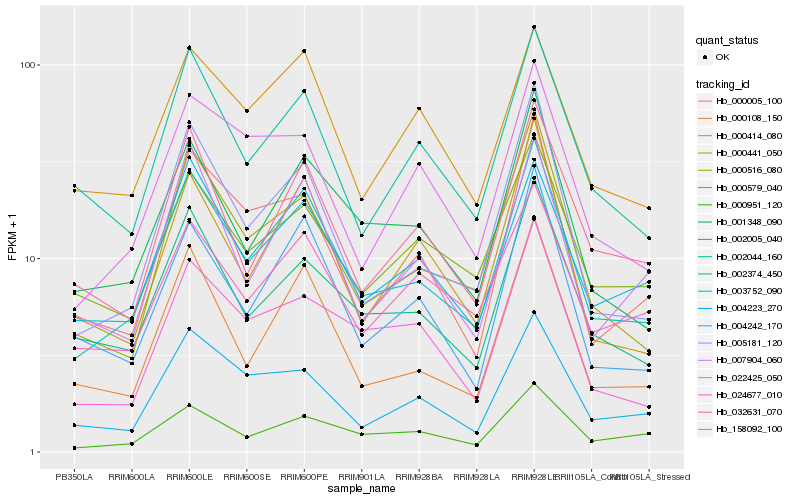
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_450 |
0.0 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001348_090 |
0.0904832445 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000414_080 |
0.0946631747 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000441_050 |
0.0959499811 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 5 |
Hb_004242_170 |
0.0969100036 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 6 |
Hb_002044_160 |
0.097428963 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 7 |
Hb_000951_120 |
0.0989885295 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 8 |
Hb_003752_090 |
0.1002500399 |
- |
- |
chitinase, putative [Ricinus communis] |
| 9 |
Hb_032631_070 |
0.1006961397 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_158092_100 |
0.1010750173 |
- |
- |
PREDICTED: uncharacterized protein LOC105647301 [Jatropha curcas] |
| 11 |
Hb_002005_040 |
0.1011563936 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_005181_120 |
0.1013526264 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_000108_150 |
0.1026284107 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 14 |
Hb_000516_080 |
0.1039743895 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 15 |
Hb_007904_060 |
0.1063954031 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 16 |
Hb_022425_050 |
0.1065393276 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 17 |
Hb_004223_270 |
0.1072096563 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 18 |
Hb_024677_010 |
0.1086181923 |
- |
- |
superoxide dismutase [fe], putative [Ricinus communis] |
| 19 |
Hb_000579_040 |
0.1087957996 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 20 |
Hb_000005_100 |
0.1088224535 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |