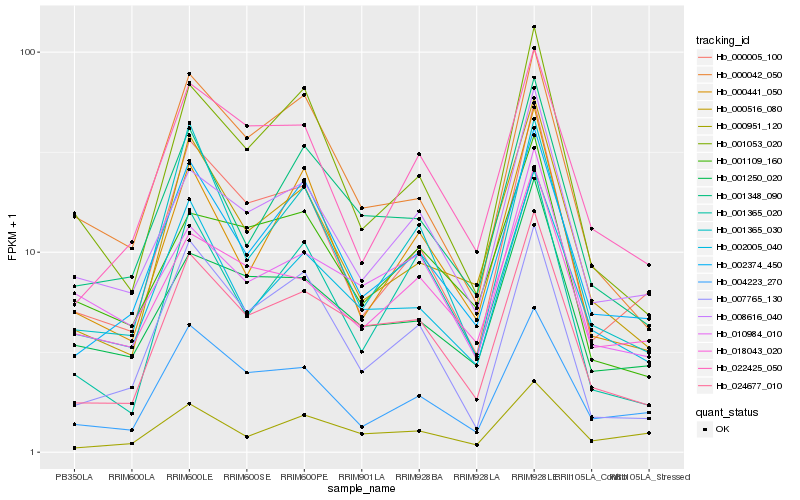
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024677_010 |
0.0 |
- |
- |
superoxide dismutase [fe], putative [Ricinus communis] |
| 2 |
Hb_001348_090 |
0.0969141248 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002374_450 |
0.1086181923 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002005_040 |
0.1138373345 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_000516_080 |
0.1196791716 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 6 |
Hb_001250_020 |
0.1198766025 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_007765_130 |
0.1213359833 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008616_040 |
0.1216887785 |
- |
- |
PREDICTED: RNA-binding protein CP33, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001053_020 |
0.123476786 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 10 |
Hb_000042_050 |
0.1262989858 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000005_100 |
0.1265524118 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_022425_050 |
0.1279246685 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 13 |
Hb_000951_120 |
0.131566592 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 14 |
Hb_001365_020 |
0.1322766143 |
- |
- |
hypothetical protein PRUPE_ppa000927mg [Prunus persica] |
| 15 |
Hb_018043_020 |
0.1325533792 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_010984_010 |
0.134318177 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_001365_030 |
0.1346103035 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 18 |
Hb_004223_270 |
0.1350989289 |
- |
- |
PREDICTED: uncharacterized protein LOC105635371 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001109_160 |
0.1363271598 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_000441_050 |
0.1386915588 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |