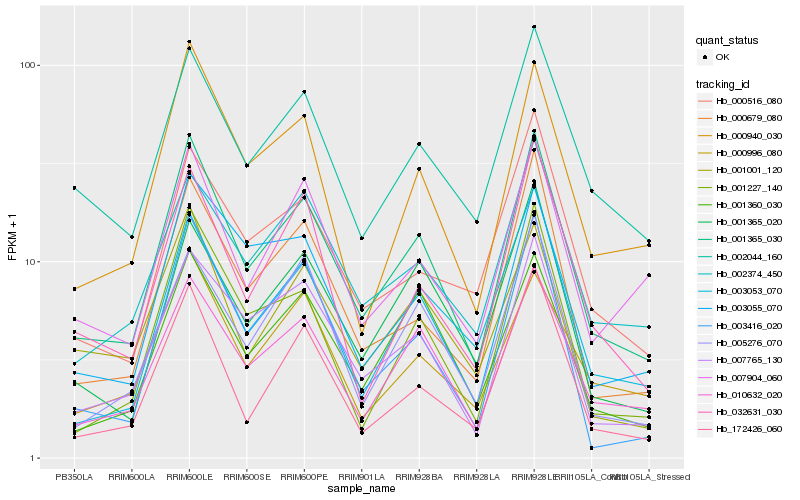
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001365_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 2 |
Hb_010632_020 |
0.0854949057 |
- |
- |
hypothetical protein POPTR_0006s185902g, partial [Populus trichocarpa] |
| 3 |
Hb_007765_130 |
0.0966303958 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000940_030 |
0.100856938 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000679_080 |
0.1053880323 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002044_160 |
0.1061069608 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 7 |
Hb_003053_070 |
0.1092650894 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002374_450 |
0.1096573719 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_007904_060 |
0.1140097719 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 10 |
Hb_001365_020 |
0.1141635199 |
- |
- |
hypothetical protein PRUPE_ppa000927mg [Prunus persica] |
| 11 |
Hb_001227_140 |
0.1144713137 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 12 |
Hb_001360_030 |
0.1147752441 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 13 |
Hb_172426_060 |
0.1153346316 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 14 |
Hb_032631_030 |
0.1158873185 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 15 |
Hb_003055_070 |
0.1164448697 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 16 |
Hb_003416_020 |
0.1169351711 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 17 |
Hb_001001_120 |
0.1175093888 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 18 |
Hb_000516_080 |
0.1177436826 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 19 |
Hb_005276_070 |
0.1182517548 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 20 |
Hb_000996_080 |
0.1190291711 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |