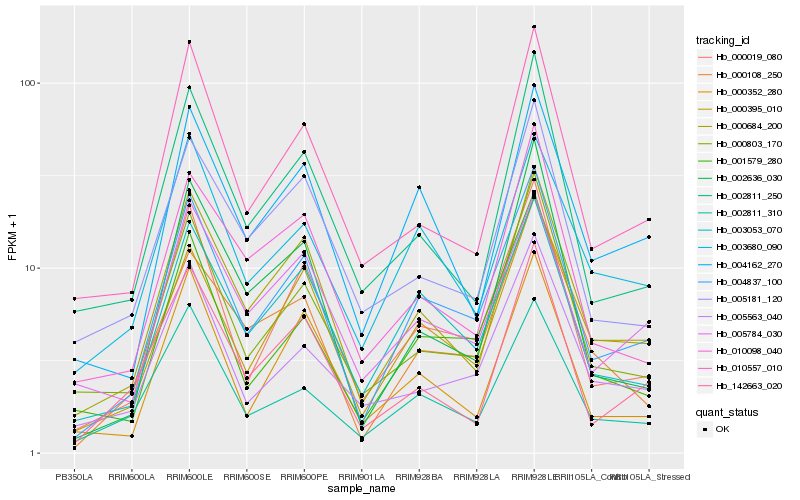
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004837_100 |
0.0 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000019_080 |
0.0790713474 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1 [Jatropha curcas] |
| 3 |
Hb_003053_070 |
0.0933955907 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000684_200 |
0.0941492254 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 5 |
Hb_000395_010 |
0.1070311411 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 6 |
Hb_001579_280 |
0.1083687579 |
- |
- |
PREDICTED: 1,4-dihydroxy-2-naphthoyl-CoA synthase, peroxisomal-like [Jatropha curcas] |
| 7 |
Hb_004162_270 |
0.1178761883 |
- |
- |
PREDICTED: superoxide dismutase [Cu-Zn], chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_000803_170 |
0.1181833226 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 9 |
Hb_000108_250 |
0.1215545044 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_010557_010 |
0.1233897259 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005784_030 |
0.1246345424 |
- |
- |
PREDICTED: uncharacterized protein LOC105645162 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002811_310 |
0.1252262558 |
- |
- |
hypothetical protein VITISV_025505 [Vitis vinifera] |
| 13 |
Hb_000352_280 |
0.1267895985 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 14 |
Hb_003680_090 |
0.1306309256 |
- |
- |
PREDICTED: threonine dehydratase biosynthetic, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005563_040 |
0.1323193433 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002811_250 |
0.1328526754 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_010098_040 |
0.1333660466 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 18 |
Hb_002636_030 |
0.1352348573 |
- |
- |
hypothetical protein JCGZ_11800 [Jatropha curcas] |
| 19 |
Hb_142663_020 |
0.1354530022 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 20 |
Hb_005181_120 |
0.1400453471 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |