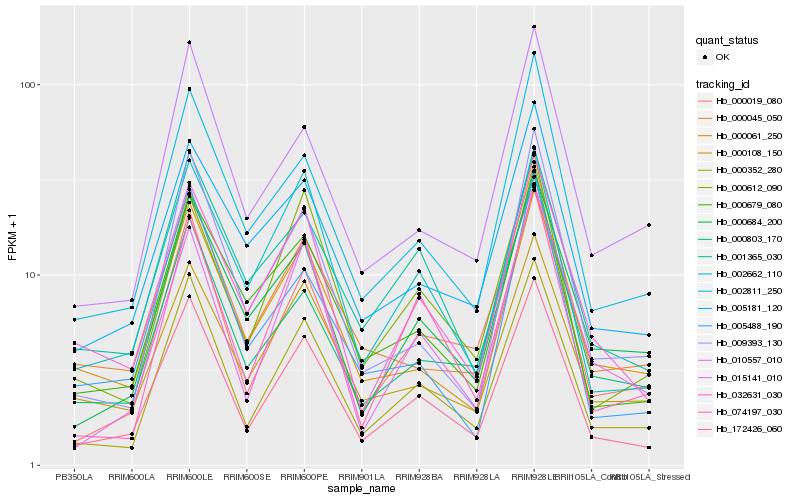
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172426_060 |
0.0 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 2 |
Hb_000352_280 |
0.0561805365 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 3 |
Hb_002811_250 |
0.0942293268 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000019_080 |
0.0945825941 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1 [Jatropha curcas] |
| 5 |
Hb_032631_030 |
0.0969205797 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 6 |
Hb_000679_080 |
0.103771401 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_015141_010 |
0.1051637416 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 8 |
Hb_074197_030 |
0.106749644 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000684_200 |
0.1096814242 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 10 |
Hb_002662_110 |
0.1100234761 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 11 |
Hb_009393_130 |
0.11196738 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 12 |
Hb_000612_090 |
0.1127166019 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 13 |
Hb_000045_050 |
0.1143626587 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000108_150 |
0.1146026237 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 15 |
Hb_001365_030 |
0.1153346316 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 16 |
Hb_010557_010 |
0.1154818181 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_005181_120 |
0.1162844683 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000061_250 |
0.1170622734 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 19 |
Hb_000803_170 |
0.1196184358 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 20 |
Hb_005488_190 |
0.1196250909 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |