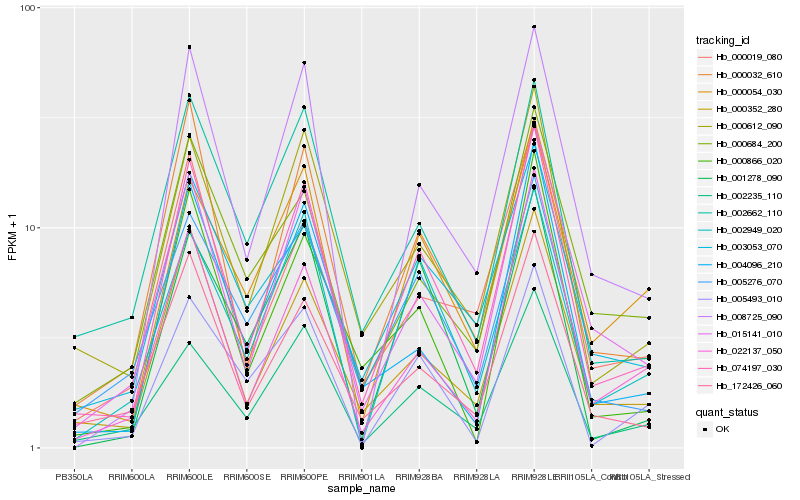
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074197_030 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000352_280 |
0.0898180173 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 3 |
Hb_000612_090 |
0.0946948286 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 4 |
Hb_015141_010 |
0.10109606 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 5 |
Hb_002235_110 |
0.106099381 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 6 |
Hb_172426_060 |
0.106749644 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 7 |
Hb_008725_090 |
0.1108360356 |
- |
- |
PREDICTED: uncharacterized protein LOC105642581 [Jatropha curcas] |
| 8 |
Hb_000032_610 |
0.1197995783 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase I, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_005276_070 |
0.120520221 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 10 |
Hb_000866_020 |
0.1228019744 |
- |
- |
hypothetical protein B456_004G042000 [Gossypium raimondii] |
| 11 |
Hb_002662_110 |
0.1228028497 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 12 |
Hb_002949_020 |
0.1232683429 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0002s23650g [Populus trichocarpa] |
| 13 |
Hb_000054_030 |
0.123891417 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 14 |
Hb_003053_070 |
0.1250359568 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000019_080 |
0.1277555462 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1 [Jatropha curcas] |
| 16 |
Hb_004096_210 |
0.128658873 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 17 |
Hb_022137_050 |
0.1291170315 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 18 |
Hb_005493_010 |
0.1327722841 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_000684_200 |
0.1339906585 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 20 |
Hb_001278_090 |
0.1368552418 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |