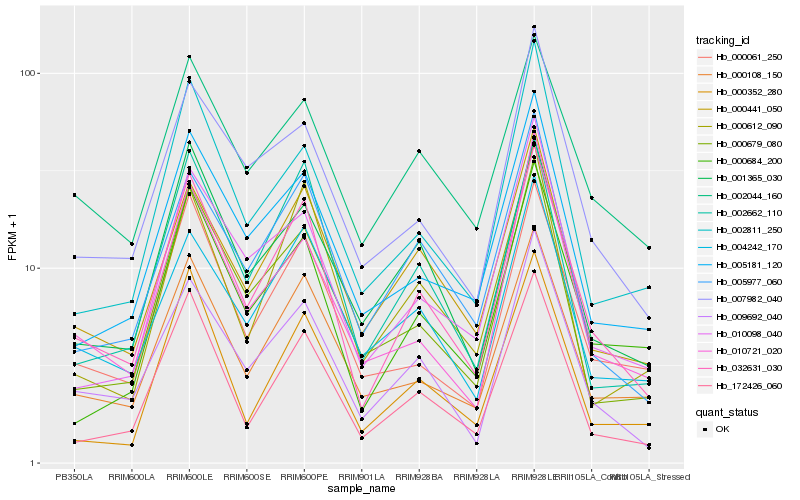
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032631_030 |
0.0 |
- |
- |
phosphatidic acid phosphatase, putative [Ricinus communis] |
| 2 |
Hb_009692_040 |
0.0905312841 |
- |
- |
PREDICTED: peptide chain release factor PrfB3, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000108_150 |
0.0955465543 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 4 |
Hb_172426_060 |
0.0969205797 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 5 |
Hb_000061_250 |
0.1007745129 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 6 |
Hb_002662_110 |
0.1031064272 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 7 |
Hb_000441_050 |
0.1051254812 |
- |
- |
PREDICTED: 187-kDa microtubule-associated protein AIR9 [Jatropha curcas] |
| 8 |
Hb_000684_200 |
0.1103785116 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 9 |
Hb_005977_060 |
0.1110657691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005181_120 |
0.1114644156 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000679_080 |
0.1133399482 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007982_040 |
0.113715579 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 13 |
Hb_004242_170 |
0.1137598184 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 14 |
Hb_001365_030 |
0.1158873185 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 15 |
Hb_002044_160 |
0.1179605432 |
- |
- |
PREDICTED: glyoxylate/succinic semialdehyde reductase 1 [Jatropha curcas] |
| 16 |
Hb_000352_280 |
0.1183549952 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 17 |
Hb_002811_250 |
0.1220491812 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_010098_040 |
0.1223388144 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 19 |
Hb_000612_090 |
0.1248237816 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 20 |
Hb_010721_020 |
0.1251296999 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |